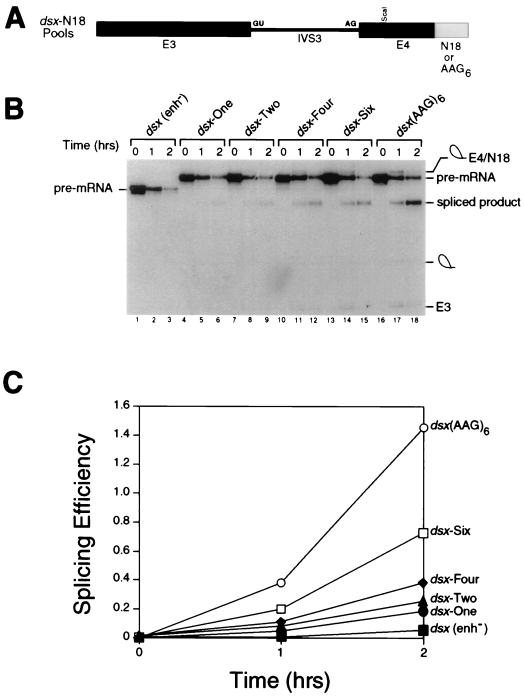
FIG. 2.
Evolution of the dsx-N18 pool. (A) The dsx-N18 and dsx-(AAG)6 constructs are shown schematically. Exon 3, intron 3, exon 4, and the enhancer(s) are indicated by E3, IVS3, E4, and N18 or AAG6, respectively. The 5′ and 3′ splice sites are indicated by GU and AG, respectively. (B) Kinetic analysis showing in vitro splicing assays performed with HeLa cell nuclear extracts and uniformly labeled pre-mRNA splicing substrates comprising the total pool of dsx-N18 pre-mRNAs after various rounds of the selection (rounds 1, 2, 4, and 6 are shown in lanes 4 to 6, 7 to 9, 10 to 12, and 13 to 15, respectively). The negative control pre-mRNA (lanes 1 to 3) is an dsx pre-mRNA lacking an enhancer [dsx(enh−)]. The positive control pre-mRNA (lanes 16 to 18) is a dsx pre-mRNA activated by six consecutive copies of a multimerized AAG trinucleotide splicing enhancer (modeled after a synthetic polypurine splicing enhancer in reference 66) that is otherwise isogenic to the dsx-N18 construct. In the kinetic analysis shown, the reaction mixtures were incubated for the number of hours indicated at the top, and positions of the precursors, intermediates, and products of the splicing reaction are indicated to the left and right. The RNAs were analyzed on a 10% denaturing gel in order to resolve the lariat-exon 4 intermediate from the spliced product. (C) Quantitation of the in vitro splicing reactions in panel B. The splicing efficiency (ratio of spliced product to precursor) is calculated from quantitation of individual bands after subtraction of background using a BAS2000 phosphorimager.