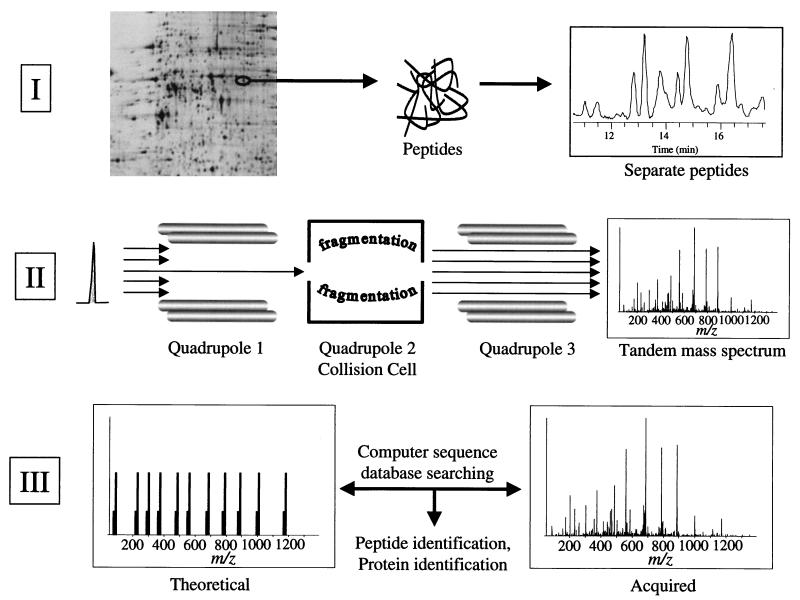
FIG. 1.
Schematic illustration of proteome analysis by 2DE and mass spectrometry. In part I, proteins are separated by 2DE, stained spots are excised and subjected to in-gel digestion with trypsin, and the resulting peptides are separated by on-line capillary high-performance liquid chromatography. In part II, a peptide is shown eluting from the column in part I. The peptide is ionized by electrospray ionization and enters the mass spectrometer. The mass of the ionized peptide is detected, and the first quadrupole mass filter allows only the specific mass-to-charge ratio of the selected peptide ion to pass into the collision cell. In the collision cell, the energized, ionized peptides collide with neutral argon gas molecules. Fragmentation of the peptide is essentially random but occurs mainly at the peptide bonds, resulting in smaller peptides of differing lengths (masses). These peptide fragments are detected as a tandem mass (MS/MS) spectrum in the third quadrupole mass filter where two ion series are recorded simultaneously, one each from sequencing inward from the N and C termini of the peptide, respectively. In part III, the MS/MS spectrum from the selected, ionized peptide is compared to predicted tandem mass spectra computer generated from a sequence database. Provided that the peptide sequence exists in the database, the peptide and, by association, the protein from which the peptide was derived can be identified. Unambiguous protein identification is attained in a single analysis because multiple peptides are identified as being derived from the same protein.