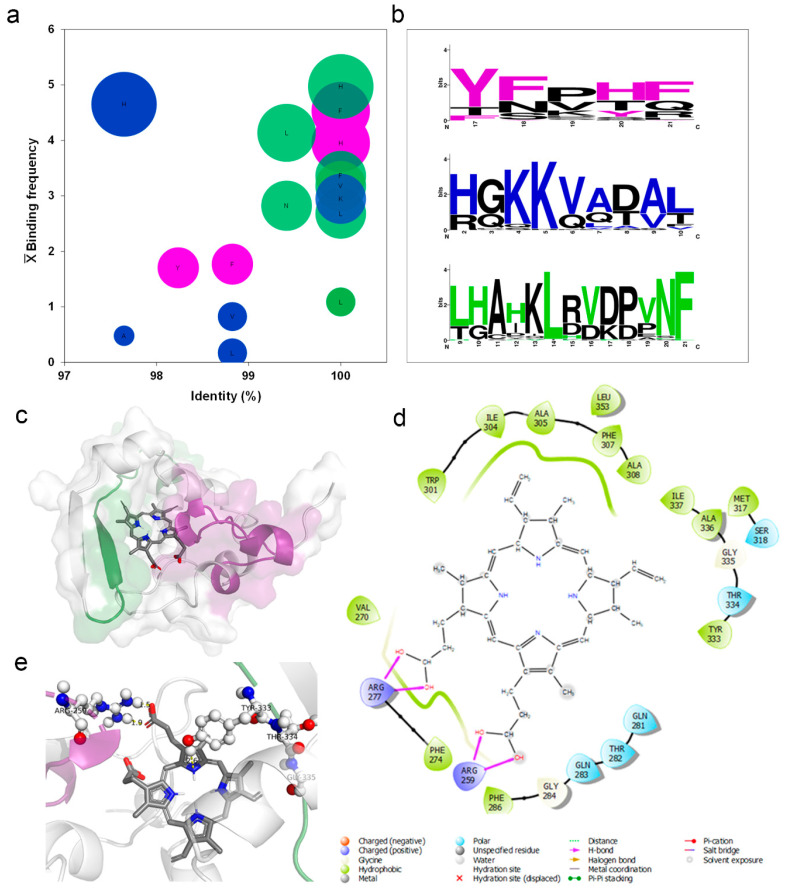
Figure 4.
Heme-binding motifs were identified by an in silico analysis. (a) The heme-binding motifs for 68 Hb structures deposited in the PDB data bank were mapped to define the frequency of residues occurrence (Binding frequency) and degree of identity (%). The ball size is relative to the number of connections (5 to 1), displayed from biggest to smallest in size. (b) Three binding motifs were identified in the N protein of SARS-CoV-2 by an analysis in the MEME-Suite server of Hb amino acid sequence obtained from UniProt sever. Colored amino acids related to residues in Hb that interact with heme. Black residues form part of the motif without binding heme. The size of the amino acid relates to the number of occurrences (bits). Molecular docking of SARS CoV-2 N protein with human PpIX. (c) Binding position of PpIX (sticks) indicating the orientation of binding with N protein (surface) to predicted motifs: purple = motif one and green = motif 3. (d) Two-dimensional (2D) representation indicating types of bonds that occur in N protein pocket bounded with PpIX. (e) Three-dimensional (3D) model showing nucleoprotein amino acid residues (ball and sticks) that compose motifs 1 and 3 coordinating the binding with the PpIX (bars).