Fig. 2. A forward-genetic, in vitro screen yields HS mutants in Msm.
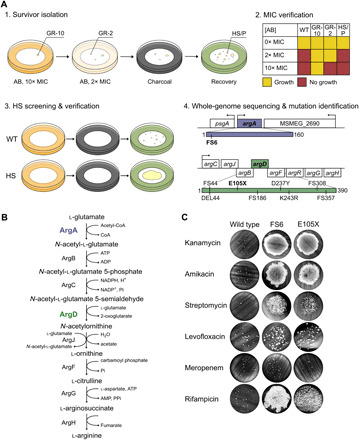
(A) HS mutants are isolated in vitro in four steps. (1) Survivor isolation. AB, antibiotic. GR-10, genetic MIC-shifted resister with ΔMIC > 10-fold. GR-2, genetic MIC-shifted resister with ΔMIC between 2- and 10-fold. HS, high-survival mutant. P, WT persister. (2) MIC verification. (3) HS screening and verification. (4) Whole-genome sequencing and mutation identification. We identified one Msm mutant with a mutation in argA (purple) and eight with mutations in argD (green), all isolated on kanamycin. Mutations annotated by their effect on the protein product. FS, frameshift. DEL, deletion (DEL44 is missing amino acids 44 to 51). (B) ArgA (purple) and ArgD (green) are members of the arginine biosynthesis pathway. CoA, coenzyme A. ATP, adenosine triphosphate. ADP, adenosine diphosphate. NADP+, nicotinamide adenine dinucleotide phosphate. NADPH, reduced form of NADP+. AMP, adenosine monophosphate. Pi, inorganic phosphate; PPi, inorganic pyrophosphate. (C) Survival of ArgA-FS6 and ArgD-E105X upon exposure to 10× the MIC of the aminoglycosides kanamycin (KAN), amikacin (AMK), and streptomycin (STR); levofloxacin (LVX); meropenem (MEM); or rifampicin (RIF). Representative of 2 to 36 independent experiments (FS6: KAN 13, AMK 2, STR 2, LVX 2, MEM 2, RIF 2; E105X: KAN 36, AMK 3, STR 3, LVX 2, MEM 2, RIF 2). See also fig. S1 and table S2.
