Fig. 6. Analysis of HIF- and PADI4-dependent gene expression in SUM159 cells by RNA-seq.
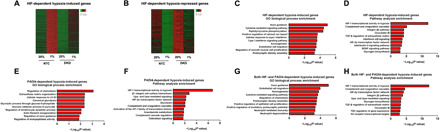
(A and B) Heat maps were prepared showing relative mRNA levels in NTC and HIF-DKD cells exposed to 20 or 1% O2 for 24 hours for 1307 HIF-dependent hypoxia-induced genes (A) and 817 HIF-dependent hypoxia-repressed genes (B). (C to H) HIF-dependent hypoxia-induced genes (C and D), PADI4-dependent hypoxia-induced genes (E and F), and HIF- and PADI4-dependent hypoxia-induced genes (G and H) were subjected to Gene Ontology (GO) biological process analysis (C, E, and G) and gene pathway enrichment analysis (D, F, and H). The top 10 categories for each analysis are shown.
