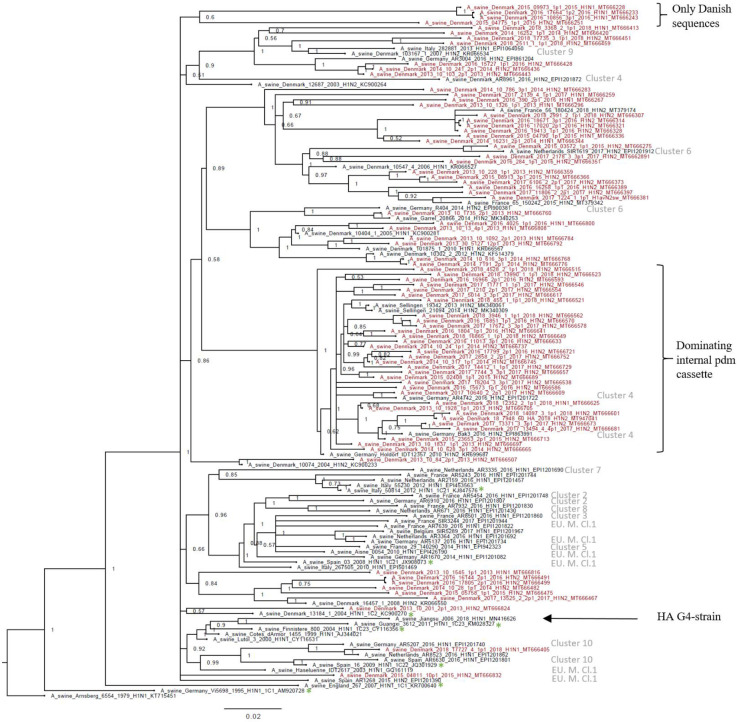
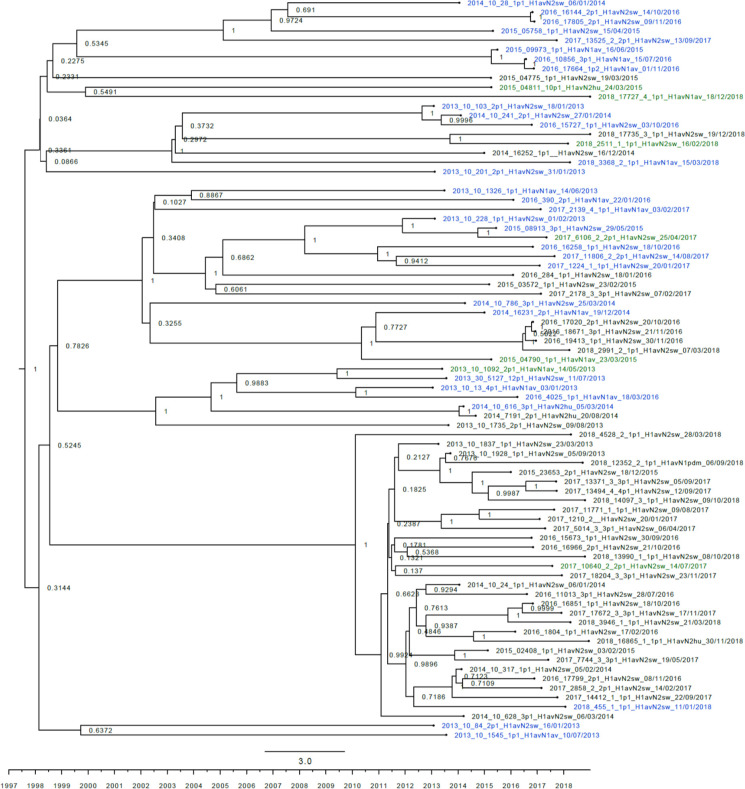
Figure 2. Bayesian phylogenetic tree of the Danish hemagglutinin (HA) nucleotide sequences of H1av origin from 2011 to 2018 and HA reference sequences of H1av origin.
The Danish HA sequences of H1av origin obtained in this study are marked in red, whereas the reference sequences of H1av are marked in black. The sequences belonging to the 10 recently defined European cluster (Henritzi et al., 2020) are highlighted with the name of the specific cluster in gray color. Two clusters are highlighted with brackets; one containing only Danish sequences and one containing H1av sequences derived from H1avNx viruses containing mainly and internal gene cassette of H1N1pdm09 origin. The reference sequences of IRD used to define the 1.C.1 (1C1) and 1.C.2 (1C2) clades are marked with a green *. Node labels represent posterior probabilities. All sequences are named according to the influenza nomenclature, and the accession number is given as a suffix for each sequence. A_Arnsberg_6554_1979_H1N1_KT715451 was used as an outgroup.