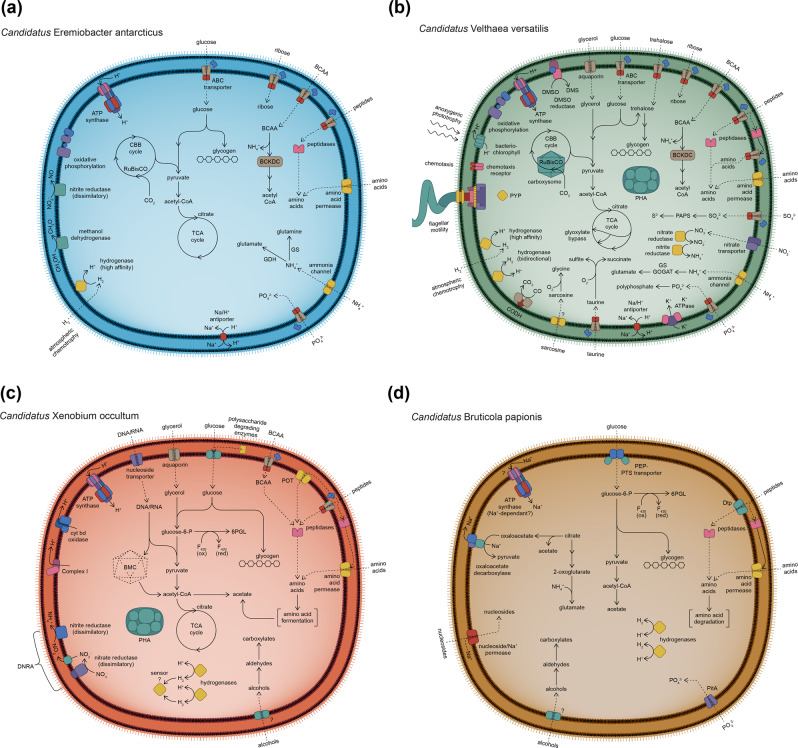
Fig. 4. Illustrations depicting the predicted diverse metabolic capacities within phylum Candidatus Eremiobacterota.
Class Candidatus Eremiobacteria a order Candidatus Eremiobacterales, represented by Candidatus Eremiobacter antarcticus sp. nov.; b order Candidatus Baltobacterales, represented by Candidatus Velthaea versatilis gen. et. sp. nov.; and class Candidatus Xenobia class. nov., represented by c Candidatus Xenobium occultum gen. et. sp. nov.; and d Candidatus Bruticola papionis gen. et. sp. nov. All cells are shown as diderm, based on the presence of lipopolysaccharide biosynthesis genes across Ca. Eremiobacterota (Supplementary Table S5). All cells are shown as coccoid, based on observed cell morphology of Antarctic Ca. Eremiobacteria cells, even though the rod-cell shape determinant MreB was encoded across both classes (Supplementary Table S5). ABC ATP-binding cassette, BCAA branched-chain amino acids, BCKDC branched-chain 2-oxoacid dehydrogenase complex, BMC bacterial microcompartment, CBB cycle Calvin–Benson–Bassham cycle, CoA coenzyme A, CODH carbon monoxide dehydrogenase, cyt bd cytochrome bd, DMS dimethylsulfide, DMSO dimethylsulfoxide, DNRA dissimilatory nitrate reduction to ammonia, Dtp di-/tripeptide transporter, F420 coenzyme F420 (8-hydroxy-5-deazaflavin), GDH glutamate dehydrogenase, GS glutamine synthetase, GOGAT glutamate synthase, ox oxidized, PAPS 3′-phosphoadenosine-5′-phosphosulfate, PEP phosphoenolpyruvate, PHA polyhydroxyalkanoate (storage product), PitA inorganic phosphate transporter, POT proton-dependent oligopeptide transporter, PTS phosphotransferase, PYP photoactive yellow protein, red reduced, RuBisCO ribulose-bisphosphate carboxylase/oxygenase, TCA cycle tricarboxylic acid cycle, 6PGL 6-phosphogluconolactone.