Fig. 6. Streptococcal hydrogen peroxide inhibits gingipain activity and prevents P. gingivalis-induced Notch signaling.
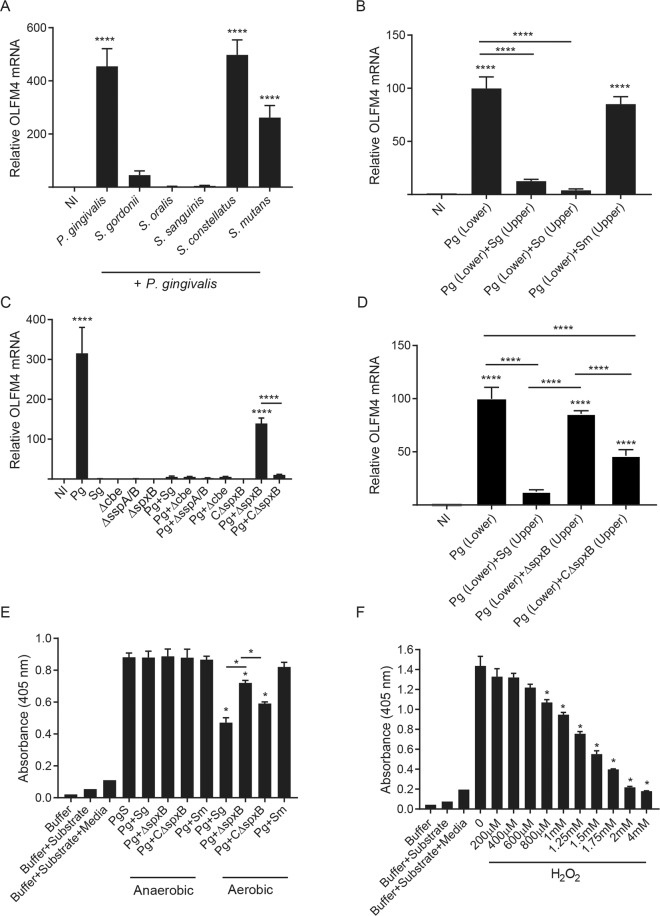
A qRT-PCR of OLFM4 mRNA in TIGK cells left uninfected (NI) or challenged with P. gingivalis alone or in combination with S. gordonii, S. oralis, S. sanguinis, S. constellatus, or S. mutans for a total MOI of 100. B TIGK cells were grown in the lower chamber of a transwell plate and challenged with P. gingivalis (Pg) or left uninfected (NI). S. gordonii (Sg), S. oralis (So) or S. mutans (Sm) were added to the upper chamber where indicated, and OLFM4 mRNA was measured by qRT-PCR. C OLFM4 mRNA levels in TIGKs uninfected (NI), challenged with P. gingivalis (Pg), S. gordonii WT or S. gordonii mutant strains alone or together in equal numbers for a total MOI 100. D TIGK cells were grown in the lower chamber of a transwell plate and challenged with P. gingivalis (Pg) or left uninfected (NI). S. gordonii WT (Sg), S. gordonii SpxB mutant (ΔspxB), or complemented SpxB mutant (CΔspxB) were added to the upper chamber where indicated, and OLFM4 mRNA was measured by qRT-PCR. Data are means with SEM. ****P < 0.001 compared to NI unless indicated. E P. gingivalis supernatant (PgS) was incubated with S. gordonii WT (Sg), SpxB mutant (ΔspxB), complemented SpxB mutant (CΔspxB) or S. mutans (Sm), cultured aerobically or anaerobically. Activity of RgpA/B was measured through cleavage of L-BAPNA chromogenic substrate. Data are means with SD and representative of three biological replicates. *p < 0.05, compared to PgS unless indicated. F P. gingivalis culture supernatant was reacted with the indicated concentrations of H2O2, and activity of RgpA/B was measured through cleavage of L-BAPNA chromogenic substrate. Data are means with SD and representative of three biological replicates. *p < 0.05, compared to P. gingivalis supernatant alone (0).