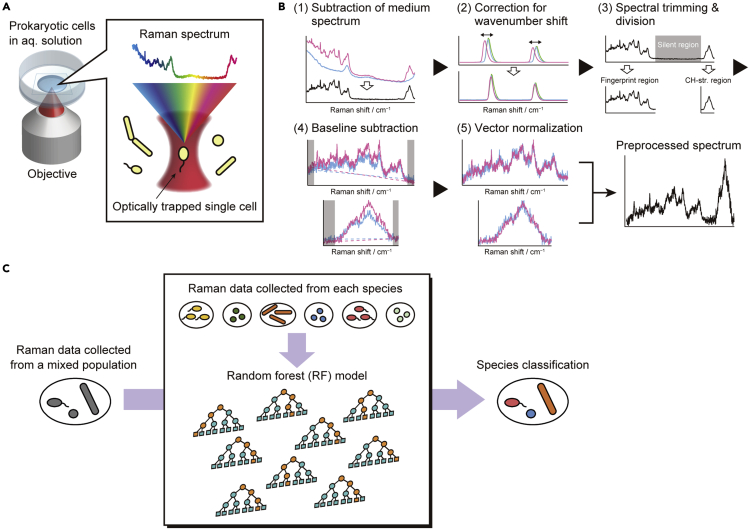
Figure 1.
Workflow of the prokaryotic classification method developed in this study, using single-cell Raman microspectroscopy and RF algorithm
(A) Acquisition of the Raman spectra of single prokaryotic cells in aqueous solution (PBS) using an optical tweezer achieved by the same laser beam at 632.8 nm as that used for the Raman excitation (see STAR Methods).
(B) Preprocessing of the measured single-cell Raman spectra: (1) subtraction of the PBS spectrum (blue line) from the cell + PBS spectrum (red line), yielding a difference spectrum (black line); (2) correction for a wavenumber shift that typically occurs among the data taken on different days (red, green, and blue lines); (3) Deletion of the so-called silent region of the Raman spectrum (gray area) and division of the spectrum into the two parts: the fingerprint and CH-stretching regions; (4) subtraction of a linear baseline (dashed lines) that is determined from the edge regions (gray areas); and (5) vector normalization, in which each Raman intensity is divided by the square root of the sum of the squared intensities of the spectrum. The preprocessed spectra in the fingerprint and CH-stretching regions are finally combined.
(C) RF model construction using the preprocessed single-cell Raman data obtained from each prokaryotic species and application to species classification in a mixed population. Training was done exclusively using the Raman spectral data collected from the pure populations of the six species.