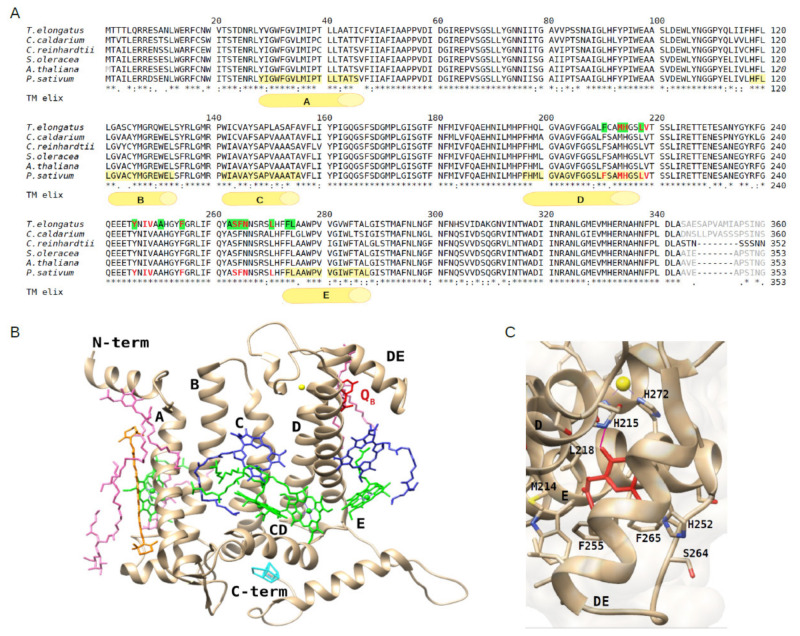
Figure 1.
Alignment of the D1 protein sequences retrieved from high-resolution structures (chain A of each PDB entry) of representatives of cyanobacteria, algae and plants and view of the D1 protein structure and the QB binding site of P. sativum. (A) Alignment of D1 sequences from the red alga Cyanidium caldarium (UniProtKB O19895, PDB: 4YUU at 2.7 Å), green alga Chlamydomonas reinhardtii (UniProtKB P07753, PDB: 6KAC at 2.7 Å) and higher plants Spinacia oleracea (UniProtKB P69560, PDB: 3JCU at 3.2 Å), Arabidopsis thaliana (UniProtKB P83755, PDB: 5MDX at 5.3 Å) and Pisum sativum (UniProtKB P06585, PDB: 5XNL, at 2.7 Å) along with that from the cyanobacterium Termosynechococcus elongatus (UniProtKB P0A444, PDB: 4V82, at 3.2 Å). In the latter, the amino acids of the QB niche involved in the terbutryn binding are highlighted in green. Amino acids surrounding the plastoquinone QB head are colored in red in the sequences from T. elongatus and P. sativum. Amino acids not resolved in the structures are indicated in grey. The five transmembrane α-helices of the D1 protein of P. sativum, indicated by letters A–E, are highlighted in yellow in the corresponding sequence. (B) View of the D1 protein and cofactors from the structure of P. sativum (PDB: 5XNL, chain A). Plastoquinone QB in red, non-heme Fe in yellow, Chls in green, Pheo molecules in blue, β-carotene in orange, lipids in pink, oxygen evolving complex (OEC) in cyan. (C) View of the QB binding site within the D1 protein rotated with respect to the view in panel B to better show the plastoquinone head (in red), the surrounding amino acid residues and the hydrogen bond (indicated as purple segment).