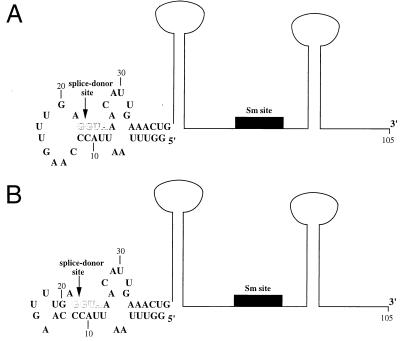
FIG. 2.
Predicted secondary structures of SL1 RNA. (A) Predicted secondary structure of SL1 RNA (6); the stem loop structure depicted for nucleotides 1 to 38 was determined by NMR (14). An outline of the rest of the structure (67 nt), generated by computer prediction (6), is indicated. The position of the 8-nt Sm binding site is represented by a solid box. The arrow indicates the position of the splice donor site; the sequence 5′ of this site comprises the leader exon. The G/GUA of the splice donor site (outlined letters) is conserved in all known SL RNAs, except in the recently identified minor SL RNAs in C. elegans (SL3, SL4, and SL5), which contain the sequence G/GUU (10, 34). In addition, the predicted base pair interactions that occur between these nucleotides and the leader are also highly conserved in SL RNAs (6). (B) An alternative structure determined by NMR analysis for the first stem-loop of the SL1 RNA (42); an outline of the rest of the structure as predicted (6) is shown and labeled as in panel A. This structure differs from that in panel A in that the nucleotides at positions 13, 14, 19, and 20 are base paired instead of forming part of the single-stranded loop.