Figure 1.
Adenine base editors can edit the DUX4 PAS
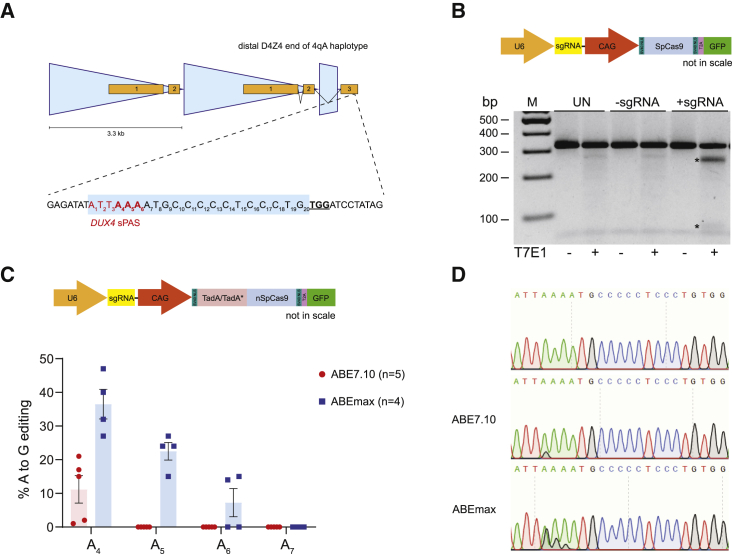
(A) Schematic representation of the distal end of the 4qA-derived D4Z4 macrosatellite repeat (each blue triangle represents one D4Z4 repeat unit) including the adjacent downstream sequence containing the polyadenylation signal of DUX4 in exon 3 (DUX4 exons are indicated by orange boxes) and zoom in on the sequence to be targeted by the adenine base editor. The sgRNA protospacer is outlined in the blue box, the PAM site for SpCas9 is underlined in bold and the DUX4 PAS sequence (ATTAAA) is in red font with adenines that can be targeted by the adenine base editor in bold. (B) Schematic map of the pX458 vector for simultaneous sgRNA and SpCas9 nuclease expression (top). Result of the T7E1 assay performed on HAP1 cells that were transfected with the pX458 vector expressing the sgRNA targeting the DUX4 PAS together with a plasmid encoding for puromycin resistance to select for transfected HAP1 cells (bottom). Untransfected cells (UN) or cells transfected with no sgRNA-containing vector (−sgRNA) served as negative control. Asterisks mark the T7E1 cleavage products. (C) Schematic map of the modified all-in-one pX458 vector encoding for the adenine base editors (top). Editing efficiency was assessed in HAP1 cells for the ABE7.10 and ABEmax versions of the adenine base editor. The A→G editing efficiency was calculated from Sanger sequencing tracks with EditR39 for each adenine in the editing window. Graph shows mean ± SEM of at least four independent biological replicates (dots). (D) Representative Sanger sequencing tracks for ABE7.10- or ABEmax-mediated editing of the DUX4 PAS used for quantification.