Figure 4.
Editing of the DUX4 PAS induces alternative pre-mRNA cleavage site
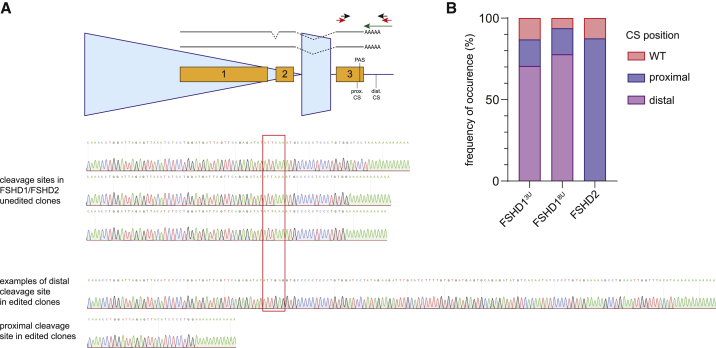
(A) Schematic of the terminal D4Z4 repeat unit with short ending (4A161S haplotype) showing the design of 3′ RACE experiment to determine the cleavage and polyadenylation site of DUX4 mRNA in the edited clones. Two known DUX4 mRNA isoforms are depicted with splicing or retention of intron 1 (top). Arrows represent primers used for oligo-dT reverse transcription (green), first PCR (red), and second nested PCR (black). The identified proximal and distal cleavage sites, for which Sanger sequencing traces are provided, are marked. Sanger sequencing tracks (bottom) show representative examples of 3′ ends of DUX4 mRNA in DUX4 PAS unedited and edited FSHD1/FSHD2 clones. The red rectangle outlines the DUX4 PAS sequence. Three different CSs were identified in unedited clones (as reported previously31), while different shifts in CSs were identified in edited clones. One representative Sanger sequencing track is shown for each CS choice. (B) Barplots representing the frequency of occurrence of different CSs identified in DUX4 PAS edited clones with respect to WT CSs from ≥4 clones for each condition.