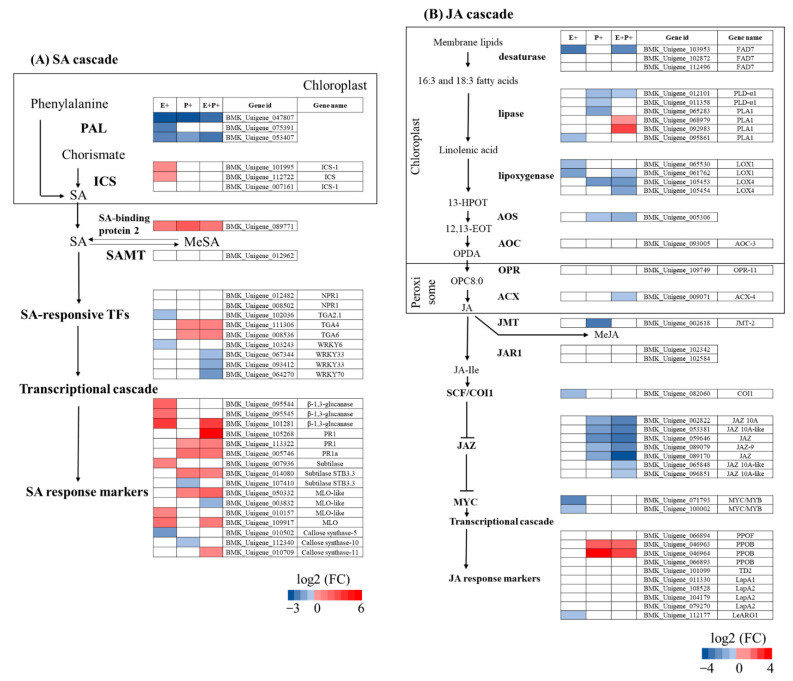
Figure 5.
Changes in expression levels of plant genes associated with the salicylic acid (SA, panel A) and jasmonic acid (JA, panel B) signalling pathways given the presence of the fungal endophyte Epichloë gansuensis (E+), the fungal pathogen Blumeria graminis (P+) and both fungi (E+P+) on Achnatherum inebrians plants. The gene expression levels were measured at four weeks after the pathogen inoculation of plants. Proteins and enzymes are shown in bold. Arrows indicate positive regulation and truncated connectors indicate the opposite. Abbreviations: PAL, phenylalanine ammonia lyase; ICS, isochorismate synthase; MeSA, methyl salicylate; SAMT, salicylate-O-methyl transferase; NPR1, nonexpressor of PR genes 1; TGA, TGACG sequence-specific binding protein; WRKY, (T)TGAC(C/T) sequence-specific binding protein; PR1, pathogenesis-related protein 1; FAD, omega-3 fatty acid desaturase; PLD, phospholipase D; PLA1, plastochron1; LOX, lipoxygenase; 13-HPOT, 13-hydroperoxyoctadeca-9,11,15-trienoic acid; AOS, allene oxide synthase; 12,13-EOT, 12,13-epoxyoctadeca-9,11,15-trienoic acid; AOC, allene oxide cyclase; OPDA, 12-oxo-phytodienoic acid; OPR, oxophytodienoic acid reductase; OPC8:0, 3-oxo-2-(cis-2’-pentenyl)cyclopentane-1-octanoic acid; ACX, acyl coenzyme-A oxidase; OPC, OPC-8:0 CoA ligase1; JMT, jasmonic acid carboxyl methyltransferase; MeJA, methyl jasmonate; JAR1, jasmonoyl--L-amino acid synthetase JAR1; JA-Ile, JA-isoleucine conjugate; COI1, coronatine-insensitive protein 1; JAZ, jasmonate ZIM domain; MYC2, bHLH transcription factor; PPOF, polyphenol oxidase F, chloroplastic; PPOB, polyphenol oxidase E, chloroplastic-like; TD2, threonine dehydratase 2 biosynthetic, chloroplastic; LapA, leucine aminopeptidasee, chloroplastic; LeARG1, arginase 1. Red and blue colors on cells indicate significant differences in gene expression levels expressed as log2 fold changes (FC) (n = 3). FC- and p-values of each gene are presented in Table S2.