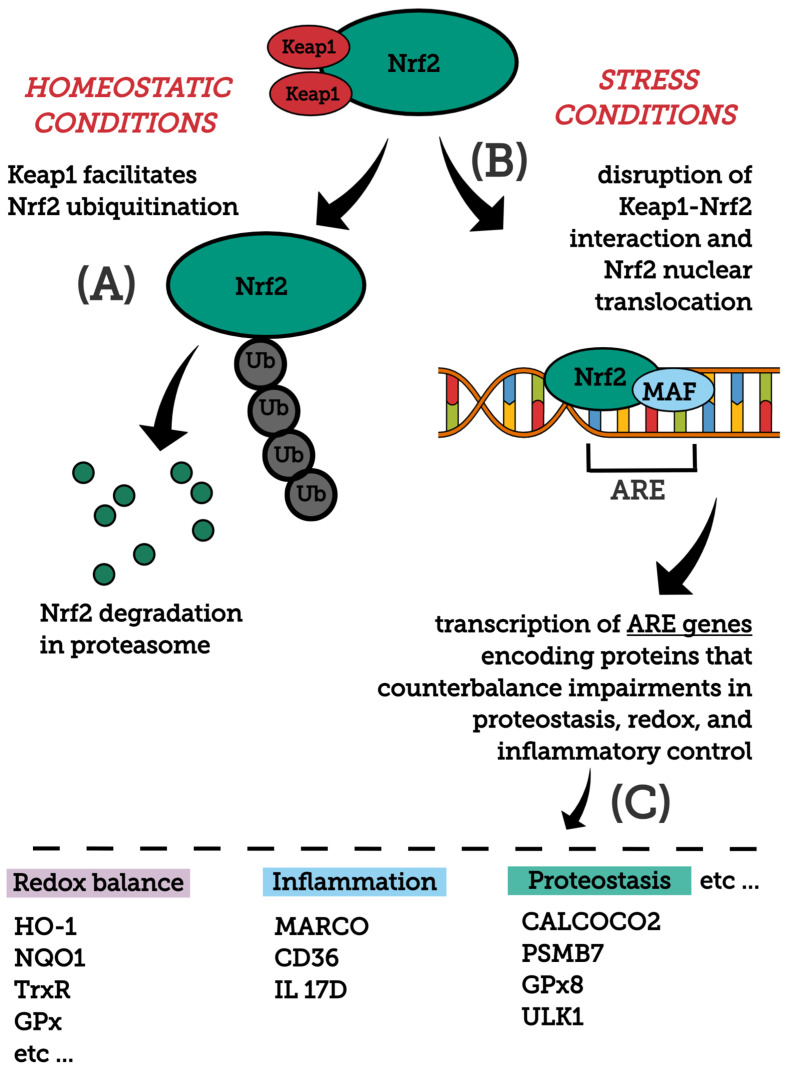
Figure 1.
General overview of the Nrf2 pathway. Major molecules and events modulating Nrf2 stability and activation, as well as main downstream protein targets and their functions are shown. (A) Nrf2 structure is composed by domains (i.e., Neh2 and Neh6) that, after redox or signaling regulation, affect Nrf2 stability. Under homeostatic conditions, Keap1 binds to Nrf2, directing this transcription factor to ubiquitination and subsequent degradation by the proteasome. Keap1 is a redox sensor that, upon oxidative thiol modification, loses its capability to repress Nrf2. Glycogen synthase kinase 3 (GSK-3)-mediated phosphorylation of Nrf2 represents an alternative mechanism, facilitating its ubiquitination and consequent degradation by the proteasome. (B) Under stress conditions (excessive accumulation of ROS, electrophilic molecules and proteotoxic stress), Nrf2-Keap1 interaction is disrupted and Nrf2 translocates into the nucleus, wherein it heterodimerizes with small Maf proteins (sMaf) and binds to an enhancer sequence termed ARE that is present in the regulatory regions of over 250 genes (ARE genes). (C) These ARE genes, whose encoded proteins participate in diverse cellular/metabolic events, display significant roles in counteracting imbalances in proteostasis, redox and inflammatory control. For a detailed review on the mechanisms mediating Nrf2 stability and activation, see [74]. CALCOCO2, calcium binding and coiled-coil domain 2; CD36, CD36 scavenger receptor; GPx, glutathione peroxidase; Gpx8, glutathione peroxidase 8; HO-1, heme oxygenase-1; IL 17D, interleukin-17D; NQO1, NADPH Quinone oxidoreductase enzyme; PSMB7, proteasome subunit b type-7; TrxR, thioredoxin reductase; ULK1, unc-51 like autophagy activating kinase 1.