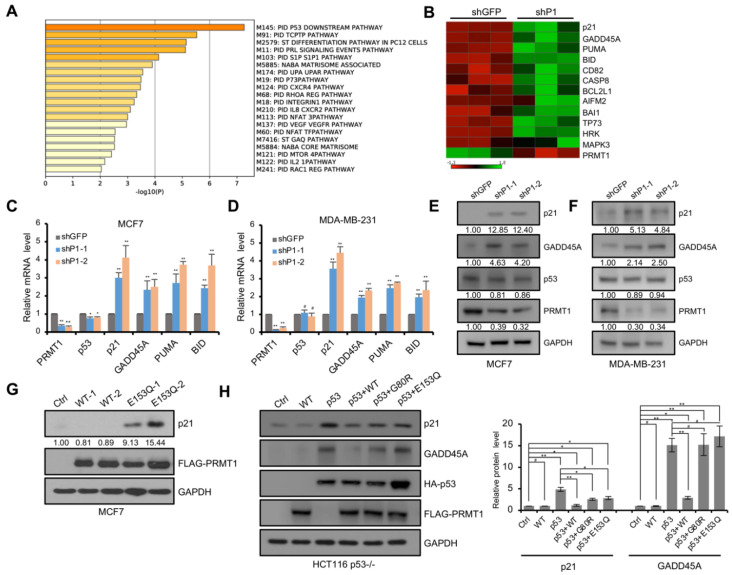
Figure 2.
PRMT1 regulated the expression of p53 targets in breast cancer cells. (A) The analysis of upregulated genes by Metascape showing the top 20 canonical pathways in shPRMT1 MDA-MB-231 cells compared with the shGFP control; p < 0.05. (B) Heatmap of p53 pathway genes in the upregulated genes (p < 0.05) analyzed with KOBAS and visualized using Matrix2png in shPRMT1 MDA-MB-231 cells. Green and red depict high and low expression levels, respectively, as indicated by the scale bar. (C–F) The expression levels of PRMT1, p53, and p53 targets were measured by RT-PCR (C,D) and Western blotting (E,F) in shPRMT1 (shP1-1 and shP1-2), and shGFP MCF7, and MDA-MB-231 cells. (G) Detection of indicated genes expression by Western blotting in MCF7 cells transfected with plasmids expressing wild-type (WT-1 and WT-2, two monoclonal cell lines) or mutated PRMT1 (E153Q-1 and E153Q-2, two monoclonal cell lines). (H) Western blotting analysis of the expression of indicated proteins in HCT116 p53−/− cells transfected with PRMT1 (WT), p53, p53 and PRMT1-WT (p53 + WT), or p53 and PRMT1-mutants (p53 + G80R and p53 + E153Q). Quantification shown on the right. The results were presented as mean ± SD from three independent experiments. #, p > 0.05; *, p < 0.05; **, p < 0.01.