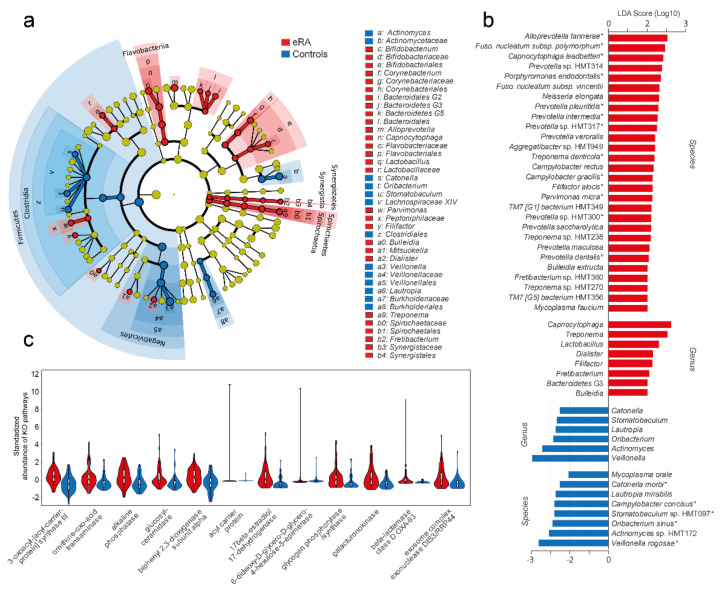
Figure 3.
Bacteria (LEfSe) and predicted functions in early onset RA versus healthy subjects. (a) Cladogram based on the phylum, class, order, family, genus and species abundances of the 340 bacterial species. (b) Bar graph showing species and genera with an LDA score > 2.0. A star (*) by the species indicates that it was detected as influential in the OPLS-DA model too (Figure 2c). (c) Violin plots with internal box plots of predicted upregulated microbiota functions in early-onset RA subjects. Using the PICRUSt algorithm, genes and gene families were predicted (KEGG orthologue) based on their 16S rRNA gene sequences. Twelve KEGG orthologues were suggested to be upregulated in early-onset RA. Mann–Whitney U test (Monte Carlo, 1000 permutations, p < 0.005) was used to evaluate group differences. All pathways are found in Table S3.