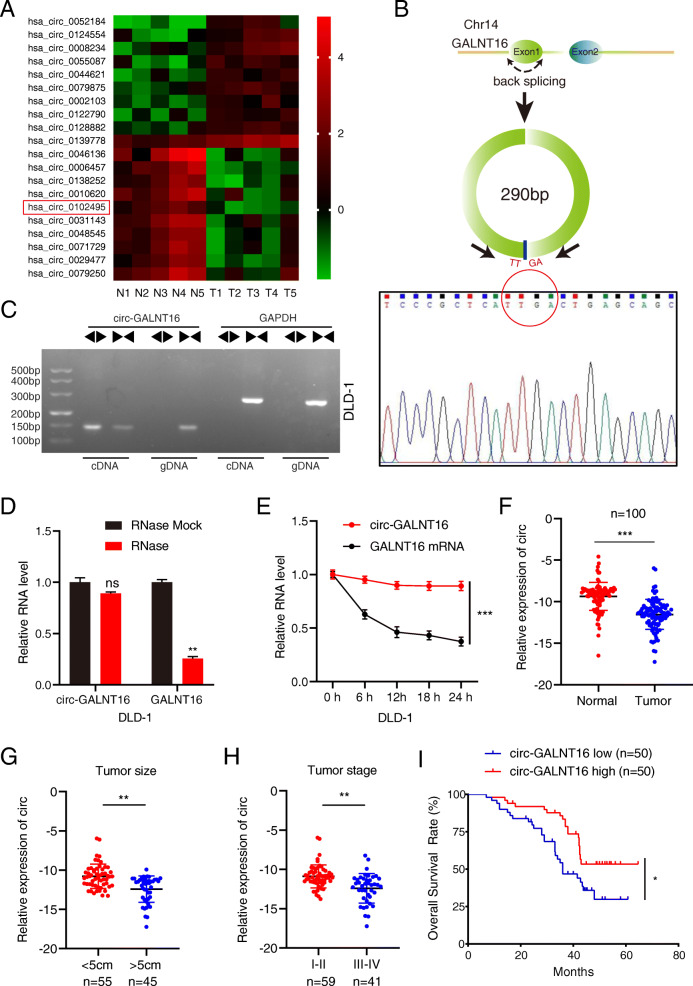
Fig. 1.
Circ-GALNT16 validation and expression in CRC cells and tissues. a Heatmap of top ten upregulated and downregulated expression circRNAs between CRC tissues and adjacent normal tissues according to the circRNA microarray. b The schematic illustration showed the back splicing of circ-GALNT16, and sanger sequence validated the splicing site. c PCR and agarose gel electrophoresis confirmed the circular formation of circ-GALNT16, using divergent and convergent primers in gDNA and cDNA of DLD-1. GAPDH was used as a negative control. d, e. Circ-GALNT16 and linear GALNT16 expression levels were detected after RNase R or actinomycin D treatment in DLD-1. f. Relative circ-GALNT16 expression in 100 CRC tissues and matched adjacent normal tissues. g-h. Relative expression of circ-GALNT16 in different tumor size and tumor stage groups. i. Kaplan-Meier plots of the overall survival of CRC patients with high (n = 50) and low (n = 50) levels of circ-GALNT16. All data are presented as the means ± SD of three independent experiments. nsp > 0.05, *p < 0.05, **p < 0.01, ***p < 0.001