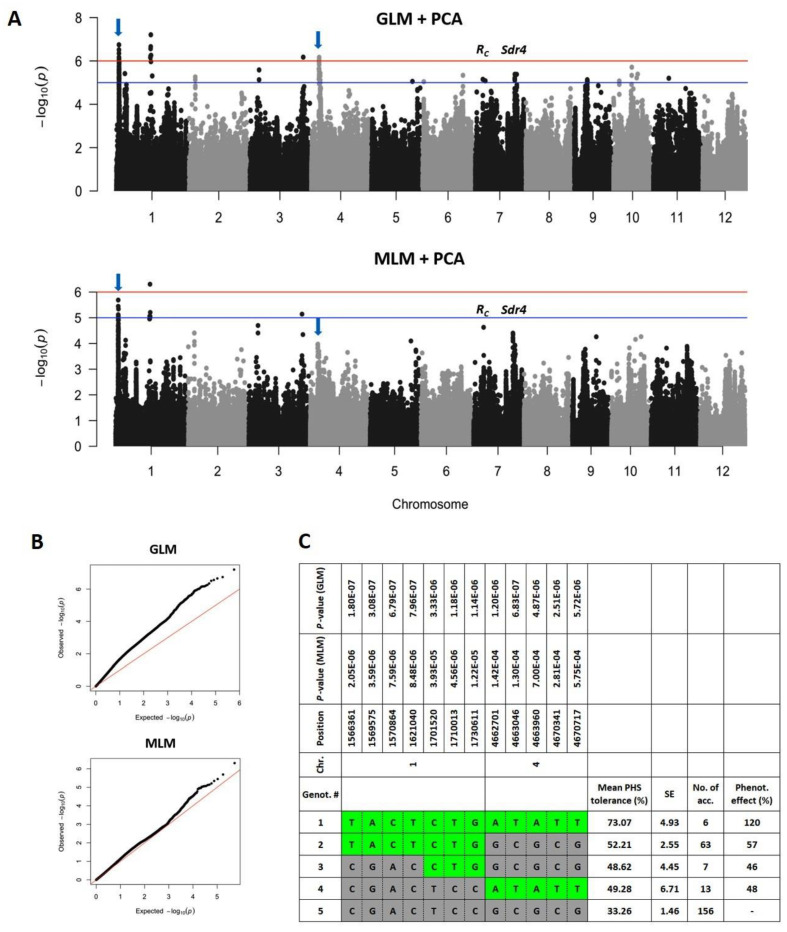
Figure 5.
Genome-wide association analysis of pre-harvest sprouting (PHS) of Japonica rice accessions: (A) Manhattan plot generated through the general linear model (GLM) with principal components and mixed linear model (MLM) with kinship matrix and principal components. Points above the blue and red threshold lines indicate significant association at −log10(p) = 5 and 6, respectively. Arrows indicate two major loci used for haplotype analysis in Figure 5C. RC and Sdr4 loci are major dormancy genes in rice; (B) quantile–quantile plots; and (C) haplotype effects on the phenotype trait.