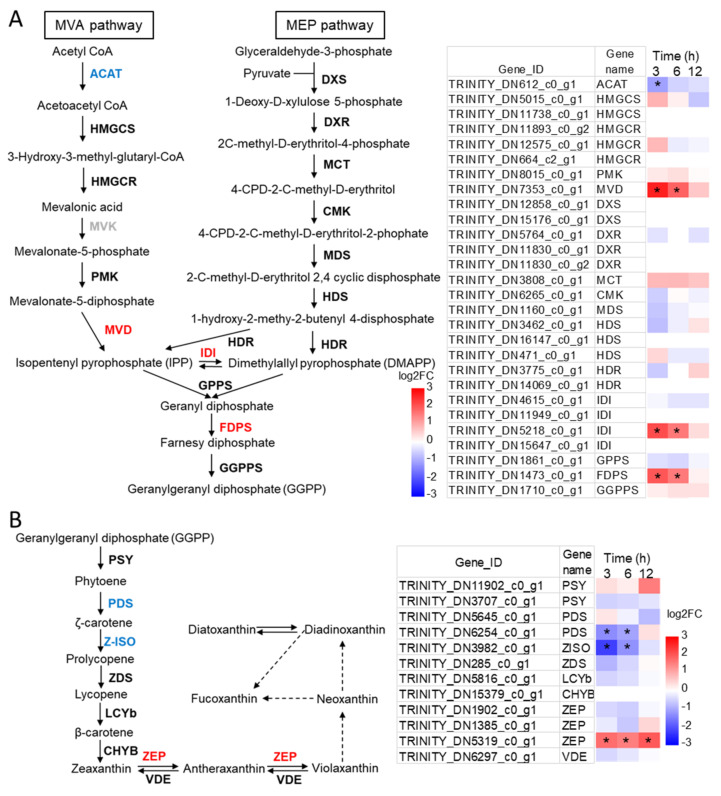
Figure 4.
Expression patterns of genes in the carotenoid biosynthesis pathway. (A) Terpenoid backbone biosynthesis, (B) Carotenoid biosynthesis. ACAT, acetyl-CoA C-acetyltransferase; HMGCS, hydroxymethylglutaryl-CoA synthase; HMGCR, hydroxymethylglutaryl-CoA reductase; PMK, phosphomevalonate kinase; MVD, diphosphomevalonate decarboxylase; DXS, 1-deoxy-D-xylulose-5-phosphate synthase; DXR, 1-deoxy-D-xylulose-5-phosphate reductoisomerase; MCT, 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase; CMK, 4-diphosphocytidyl-2-C-methyl-D-erythritol kinase; MDS, 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase; HDS, (E)-4-hydroxy-3-methylbut-2-enyl-diphosphate synthase; HDR, 4-hydroxy-3-methylbut-2-enoyl diphosphate reductase; IDI, isopentenyl-diphosphate Delta-isomerase; FDPS, farnesyl diphosphate synthase; GGPPS, geranylgeranyl diphosphate synthase; GPPS, geranyl diphosphate synthase; ZDS, zeta-carotene desaturase; PSY, phytoene synthase; PDS, phytoene desaturase; LCYb, lycopene beta-cyclase; ZEP, zeaxanthin epoxidase; VDE, violaxanthin de-epoxidase; ZISO, zeta-carotene isomerase; CHYB, beta-carotene 3-hydroxylase. * indicates the expression level in +G group was significantly differential compared with control group (|log2FC (Fold change)| ≥ 1.00 and padj < 0.05). The red color of genes indicates upregulation and blue color indicates downregulation.