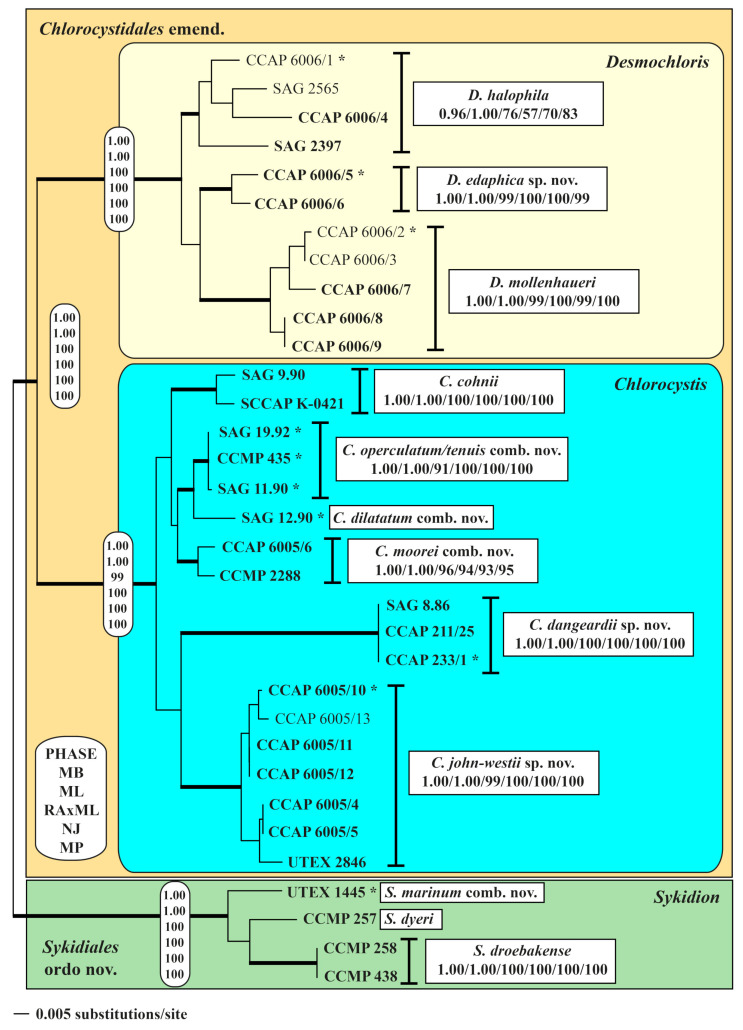
Figure 2.
Molecular phylogeny of the Chlorocystidales and Sykidiales based on SSU and ITS rDNA sequence comparisons. The phylogenetic trees shown were inferred using the maximum likelihood method based on the data sets (2410 aligned positions of 33 taxa) using PAUP 4.0a build169. For the analyses, the best model was calculated by the automated model selection tool implemented in PAUP. The setting of the best model was given as follows: SYM+I+G (base frequencies: equal; rate matrix A-C 1.1990, A-G 2.2173, A-U 1.7504, C-G 0.7565, C-U 4.3696, G-U 1.0000) with the proportion of invariable sites (I = 0.7119) and gamma shape parameter (G = 0.4903). The branches in bold are highly supported in all analyses (Bayesian values > 0.95 calculated with PHASE and MrBayes; bootstrap values > 70% calculated with PAUP using maximum likelihood, neighbor-joining, maximum parsimony, and RAxML using maximum likelihood). The authentic strains are highlighted with an asterisk behind the strain number.