Abstract
Staphylococcal infections are among the most common foodborne diseases. We performed the antibiotic susceptibility and molecular characterization of S. aureus from milk samples of dairy cows in Manhiça District. We observed a high frequency of S. aureus (41%, 58/143), in which 71% (41/58) were from commercial farms and 29% (17/58) from smallholder farms. Half of the isolates (50%, 29/58) were resistant to at least one antibiotic, with higher rates of resistance to penicillin (43%, 25/58), followed by tetracycline (16%, 9/58). Multidrug-resistant and methicillin-resistant S. aureus isolates were rare (5%, 3/58 and 3%, 2/58, respectively). The genetic diversity was low, with predominance of human-adapted strains being: ST1/CC1-t5388 (78%) and ST152-t1299 (10%), followed by ST8/CC8-t1476 (5%) and ST5/CC5-t002 (3%) and lastly, ST508/CC45-t331 and ST152-t355, with 2% each. The Panton–Valentine leukocidin (PVL) gene was detected among 14% (8/58) of the isolates, while genes encoding staphylococcal enterotoxins were scarce (3%, 2/58). Our findings revealed a high frequency of S. aureus, with high rates of resistance to the antibiotics commonly used in veterinary and human medicine. Further investigations focusing on the molecular epidemiology of S. aureus from cattle and farmers will provide detailed insights on the genetic relatedness between the strains.
Keywords: dairy cows, raw milk sample, S. aureus, antibiotic resistance, molecular typing, PVL, enterotoxins, Manhiça, Mozambique
1. Introduction
Staphylococcal foodborne disease (SFD) is one of the most common foodborne diseases and a major concern in public health programs worldwide [1]. SFD results from the consumption of contaminated foods by Staphylococcus aureus enterotoxins (SEs) that are resistant to heat treatment [2,3]. The foods that have been frequently implicated in SFD are meat and meat products, poultry and egg products and milk and dairy products [4].
In addition to causing SFD in humans, S. aureus is an opportunistic pathogen responsible for causing conditions that range from superficial skin infections to life-threatening diseases [5]. On the other hand, in animals, it is the common agent of mastitis, causing significant financial losses to dairy farms [6]. Specific strains of S. aureus can possess different virulent traits predisposing to different clinical outcomes in the host, persistence of infections and antibiotic treatment response [7]. The treatment of infections caused by S. aureus has been challenged by the emergence of multidrug-resistant (MDR) strains, including methicillin-resistant S. aureus (MRSA) [8], as the result of indiscriminate use of antibiotics both in human and veterinary medicine [9,10]. Many of the genes encoding antibiotic resistance reside on mobile genetic elements, which can be exchanged between strains from animals and humans occupying the same ecological niche [11].
Several molecular typing methods (multilocus sequence typing (MLST), spa typing, SCCmec typing and pulse-field gel electrophoresis (PFGE)) are widely used to track epidemiologically related strains, allowing us to trace the origin of SFD [12]. These typing methods have revealed evidences of humans and cattle sharing the same strains of S. aureus. In addition, of greater concern is the emergence of supposed bovine-adapted strains in the human population as well as of supposed human-adapted strains in the bovine population, including the circulation of MDR strains [13].
While antibiotic susceptibility and molecular characterization of S. aureus isolated from the food chain have been extensively studied in developed countries, the corresponding data from Africa is limited. Available data have shown S. aureus isolated from milk samples with high rates of resistance to antibiotics used in veterinary practices among African isolates [14,15], as well as the detection of indistinguishable S. aureus isolated from cattle and humans, suggesting bacterial transmission between hosts [16].
Previous studies on the antibiotic susceptibility and molecular characterization of S. aureus in Mozambique only focused on patients with community-acquired infections, nosocomial infections or in carriers [17,18,19,20,21,22], while data of S. aureus isolated from the food chain were not investigated. Therefore, in this study, we determined the antibiotic susceptibility and the clonal structure of S. aureus isolated from milk samples of dairy cows in the Manhiça District, Mozambique between April and May 2019.
2. Materials and Methods
2.1. Study Design
The study was conducted in the Manhiça District, a rural area located about 80 km north from downtown Maputo (southern Mozambique). Manhiça has a tropical climate with two seasons, a warm, rainy season between October and March, and a cool and dry season during the rest of the year [23].
Between April and May 2019, we performed a cross-sectional study in one commercial farm and among six smallholder farms in the Manhiça District during the milking season. Because of the limited number of commercial farms in the district and limited accessibility to smallholder farms, sample size calculation could not be determined.
The commercial farm and the smallholder farms were attributed an identification code for anonymization. The commercial farm included in this study commercializes milk and dairy products (cheese, yogurt) in the Manhiça District. The milk from the smallholder farms is either commercialized directly to the customers (local residents/regional travelers) or to commercial farms. The milking process in the commercial farm is either mechanized or by hand, while in the smallholder farms, only hand milking is performed.
2.2. Specimen Collection
We performed physical examination of the udder quarters of each animal for viability, and only those in the lactation phase and with at least one viable teat were included in the study; therefore, the milk sampling per animal ranged from one to four teats. Animals in the colostrum phase or in the final phase of lactation were excluded. None of the cows from this study presented any clinical symptoms related to mastitis or were under antibiotic therapy.
The udder quarters with viability were cleaned with cotton soaked in water and soap, ending by disinfection of each teat with 70% alcohol. For each teat, the first three milk jets were discarded, and an aseptic milk sample (approximately 4 mL) was collected in a sterile tube (one tube per teat), conditioned at 2–8 °C and transported to the microbiology laboratory of the Maputo Central Hospital for culture and identification.
2.3. Isolation and Identification of Staphylococcus aureus
For S. aureus isolation, each sample was homogenized by vortexing, and a sterile 10 µL plastic loop was introduced on each tube containing milk and then cultured into blood agar plates and incubated at 37 °C in a 5% CO2 atmosphere for 24 h. All positive cultures with a Gram stain compatible to staphylococci were subcultured into blood agar plates and incubated overnight as mentioned above. Distinct isolates with morphology resembling staphylococci were selected from each blood agar plate for catalase testing. Catalase positive isolates were verified by Pastorex Staph-Plus testing (Bio-Rad, Marnes-la-Coquette, France) and confirmed with the BD BBL™ Coagulase Plasma, Rabbit “tube coagulase” test (BD, Sparks, MD, USA). All the isolates identified as S. aureus by conventional microbiology were stored in Microbank™ medium and sent to the microbiology laboratory of the Centro de Investigação em Saúde de Manhiça (CISM) for molecular confirmation and downstream analysis.
2.4. Preparation of Bacterial DNA and Molecular Confirmation of S. aureus
At CISM, the isolates were retrieved from the storage and cultured on blood agar plates and incubated overnight at 37 °C in a 5% CO2 atmosphere. Afterwards, one colony was selected from the blood agar plate and inoculated into 5 mL of BD Tryptic Soy Broth followed by overnight incubation at 37 °C. Upon the incubation, 1 mL of the culture was centrifuged, and the pellets were suspended and lysed in Tris-EDTA buffer with Triton-X 100 (Aldrich Chemical Co., Milwaukee, WI, USA) and proteinase K (Sigma Chemical Co., St Louis, MO, USA) for 1 h at 56° C, followed by boiling at 100 °C for 10 min as previously described [24]. The crude DNA was diluted in distilled water (1:10) and analyzed by conventional polymerase chain reaction (PCR) for confirmation of S. aureus species through the detection of the specific nuc gene which encodes the thermostable nuclease using primers and conditions previously described [25]. The reference strains S. aureus strains ATCC® 25923TM, and Staphylococcus epidermidis ATCC® 12228TM were included as positive and negative controls, respectively. The DNA of confirmed S. aureus isolates was stored at −20 °C for further molecular characterization.
2.5. Antimicrobial Susceptibility Test
The molecularly confirmed S. aureus isolates were tested for antimicrobial susceptibility using the Kirby–Bauer disk diffusion and E-test methods, and interpreted according to the Clinical Laboratory Standards Institute (CLSI) guidelines (30th Edition) [26]. The isolates were tested against antibiotics used in veterinary medicine as well as antibiotics relevant to human medicine. The cefoxitin disk was used as a surrogate for oxacillin, for the prediction of putative methicillin-resistant S. aureus (MRSA phenotype). Inducible clindamycin resistance (ICR) was tested for all isolates resistant to erythromycin and susceptible or intermediate to clindamycin. Multidrug-resistant (MDR) phenotype was defined as resistance to ≥3 unrelated classes of antibiotics [27]. S. aureus strains ATCC® (25923 TM and 29213 TM) were used as quality controls.
2.6. Screening of Resistance Determinants
S. aureus isolates showing non-susceptibility profiles were screened for the corresponding resistance determinants (tetM, tetL, ermA, ermC, msrA, dfrG and dfrA(S1) by conventional monoplex PCR using specific primers and thermal cycling conditions (Table S1). PCR for the main resistance determinants encoding for resistance to tetracycline (tetK), cefoxitin (mecA) and penicillin (blaZ) was performed among all the isolates regardless of the observed phenotype (Table S1). The amplification products were separated through a 1.5% agarose gel stained with ethidium bromide. The 1-kb plusA 100-bp DNA ladder (Bio-Rad) was used as a molecular size marker in the gels. The S. aureus strain ATCC® 25923 TM was used as a negative control.
2.7. Detection of Virulence Genes
The S. aureus isolates were analyzed by conventional PCR using primers and conditions previously established for detection of staphylococcal enterotoxins (sea to see), toxic shock syndrome toxin (tst) [28] and Panton–Valentine leukocidin (PVL) [29] related genes. The S. aureus 111066.2 was used as a negative control [18].
2.8. Staphylococcal Protein A (spa) Typing
We performed molecular typing by amplifying and sequencing the hypervariable region of the spa gene as described elsewhere [30]. The spa types were then assigned using the Ridom Staph Type version 2.2.5 (Ridom GmbH, Würzburg, Germany).
2.9. Multilocus Sequence Typing (MLST)
MLST was performed for all the isolates, using the scheme previously described [31]. The allelic profiles, sequence types (STs) and clonal complexes were assigned using the MLST S. aureus database (https://pubmlst.org/; accessed on: 20 May 2021).
2.10. Data Analysis
The statistical analyses were performed using STATA version 14.2 (StataCorp LP, College Station, TX, USA). Categorical variables were compared using the Chi-square or Fisher’s exact test when appropriate. We deemed p-value of 0.05 or lower to be statistically significant. The antimicrobial susceptibility data were entered in a spreadsheet and analyzed through WHONET version 19.8.6 (World Health Organization, Geneva, Switzerland).
The Simpson’s index of diversity (diversity index (DI)) was used to evaluate the genetic diversity [32].
3. Results
3.1. Frequency of S. aureus Isolation
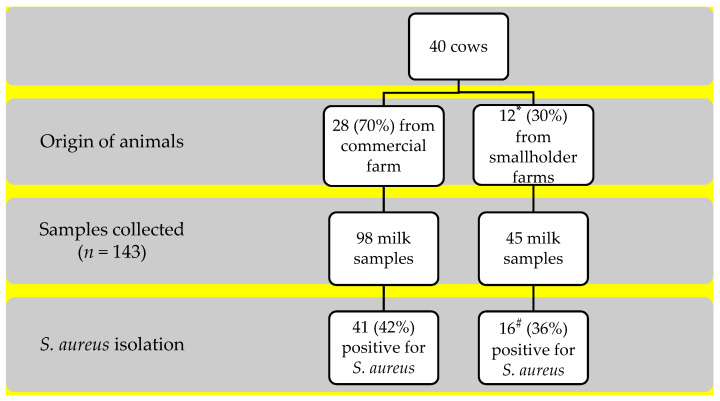
One hundred and forty-three milk samples were collected from forty cows, in which twenty-eight (70%) cows were confirmed positive to S. aureus, with a similar distribution between cows from the commercial and smallholder farms (71%, 20/28 vs. 67%, 8/12, p = 1.000). The overall frequency of positive samples for S. aureus was 40% (57/143), with two S. aureus with distinct phenotypes isolated from one sample (Figure 1). The number of positive samples ranged among the 28 cows: (i) 7 positive samples were collected from 1 teat among 7 cows; (ii) 28 samples from 2 teats among 14 cows; (iii) 18 samples from 3 teats among 6 cows; lastly, (iv) 4 samples from 4 teats in 1 cow (Table S2).
Figure 1.
Samples collected and frequency of Staphylococcus aureus isolation. * Average of two cows/smallholder farm. # In one sample two S. aureus isolates with distinct phenotypes were found.
3.2. Antibiotic Susceptibility
Half of the S. aureus isolates (50%, 29/58) were resistant to at least one antibiotic class. The MDR phenotype was rare, with three isolates (5%), showing resistance simultaneously to: (i) PEN-TCY-ERY-CD; (ii) PEN-ERY-CD; and (iii) PEN-SXT-TCY. The highest frequencies of resistance were observed for penicillin (43%), followed by tetracycline (16%), and the lowest for erythromycin/clindamycin and co-trimoxazole with 3% and 2%, respectively (Table 1). The two distinct isolates (based on colony morphology) from the same sample showed distinct resistance patterns (resistance for penicillin vs. tetracycline).
Table 1.
Antibiotic resistance of Staphylococcus aureus isolated from raw milk samples of dairy cows in southern Mozambique between April and May 2019.
| Antibiotics | Commercial Farm n = 41 (%) |
Smallholder Farms n = 17 (%) |
p | Total n = 58 (%) |
|---|---|---|---|---|
| Penicillin | 12 (29) | 13 (76) | 0.001 | 25 (43) |
| Tetracycline | 3 (7) | 6 (35) | 0.014 | 9 (16) |
| Erythromycin/Clindamycin 1 | 1 (2) | 1 (6) | 0.504 | 2 (3) |
| Co-trimoxazole | 0 | 1 (6) | NA | 1 (2) |
1 The erythromycin-resistant isolates exhibited the inducible clindamycin resistance (ICR) phenotype. NA: Not applicable.
Resistances to penicillin and to tetracycline were significantly more frequent in isolates from the smallholder farms than in the commercial farm (Table 1). All the isolates were susceptible to cefoxitin (MRSA phenotype not observed), ciprofloxacin, chloramphenicol, gentamicin, nitrofurantoin, daptomycin, linezolid and vancomycin.
3.3. Resistance Determinants
3.3.1. Resistance Determinants on Resistant Isolates
We found consistent correlation between the phenotypic resistance and its resistance determinants. Approximately half of the isolates resistant to penicillin (48%, 12/25) carried the blaZ gene encoding for beta-lactamase. Among the isolates resistant to tetracycline, all (n = 9) carried the tetK gene, while 67% (6/9) and 11% (1/9) carried the tetL and tetM genes, respectively. One of the two S. aureus isolates with the ICR phenotype simultaneously carried the mrsA and ermC genes, while the other one carried only the ermC gene; the ermA gene was not detected. The unique isolate resistant to co-trimoxazole carried the dfrG gene, while the dfrA(S1) gene was absent.
3.3.2. Resistance Determinants on Susceptible Isolates
All the isolates susceptible to penicillin (n = 33) were negative to the blaZ gene. On the other hand, 3/49 (6%) of the isolates susceptible to tetracycline were positive to the tetK gene. Two of the fifty-eight (3%) isolates phenotypically susceptible to cefoxitin harbored the mecA gene (MRSA strains).
3.4. Molecular Typing
The molecular typing revealed low diversity (DI = 0.39), in which isolates from the same spa type belonged to the same ST. We identified six spa types and five STs among the 58 S. aureus isolates: the strain ST1/CC1-t5388 (76%, n = 44) predominated, followed by ST152-t1299 (12%, n = 7), ST8/CC8-t1476 (5%, n = 3), ST5/CC5-t002 (3%, n = 2) and lastly, the ST508/CC45-t331 and ST152-t355 with 2% (n = 1) each.
Some strains were found exclusively among the smallholder farms (ST8/CC8-t1476, ST152-t355 and ST508/CC45-t331) or in the commercial farm (ST5/CC5-t002), as well as scattered in both (ST152-t1299 and ST1/CC1-t5388). S. aureus isolated from distinct teats of the same animal frequently belonged to the same clone (n = 16 animals) rather than in distinct ones (n = 5 animals), as shown in Table S2.
The two isolates from the same sample which presented distinct phenotypes (colonies morphology) belonged to distinct strains (ST1/CC1-t5388 vs. ST152-t1299). The two isolates genotypically identified as MRSA belonged to the clones ST1/CC1-t5388 and ST508/CC45-t331, and were pan-susceptible and penicillin-resistant, respectively. We did not find an association between genotypes and resistance patterns (Table 2).
Table 2.
Clonal relatedness and resistance profile of S. aureus isolated from raw milk in dairy cows in Manhiça District, southern Mozambique.
| N° Isolates Per Sector | ||||||
|---|---|---|---|---|---|---|
| Spa type (n) | MLST (n) | PVL Gene (n) | Smallholder Farms (n) | Commercial Farm (n) | Resistance Pattern (n) | Resistance Determinants (n) |
| t5388 (44) | ST1/CC1 (44) | 0 | 8 | 36 | PEN (13); PEN-TET (1); TET (1) |
blaZ (1); tetK (2); blaZ-tetK (1), mecA-tetK (1) |
| t1299 (7) | ST152 (7) | 7 | 4 | 3 | PEN (2); TET (3); PEN-TET (1), PEN-TET-ERY-CD, (1) | blaZ (1); blaZ-ermC-tetK-tetL (1); blaZ-tetK-tetL (1); tetK-tetL (2); tetK-tetM-tetL (1) |
| t1476 (3) | ST8/CC8 (3) | 0 | 3 | 0 | PEN (1); PEN-TET (1); PEN-TET-SXT (1) | blaZ (1); blaZ-tetK-tetL (1); blaZ, tetK, dfrG (1) |
| t002 (2) | ST5/CC5 (2) | 0 | 0 | 2 | PEN (2) | blaZ (2) |
| t331 (1) | ST508/CC45 (1) | 0 | 1 | 0 | PEN | blaZ-mecA-tetK |
| t355 (1) | ST152 (1) | 1 | 1 | 0 | PEN-ERY-CD | blaZ-msrA-ermC |
PEN: Penicillin; TET: Tetracycline; ERY: Erythromycin; CD: Clindamycin; SXT: Co-trimoxazole.
3.5. PVL and Staphylococcal Enterotoxins (SEs) Gene Detection
The frequency of the gene encoding for PVL toxin was low (14%, 8/58), and detected exclusively among the S. aureus isolates belonging to the ST152. The overall frequency of strains positive to SEs was low (3%, 2/58), in which one isolate (ST5/CC5-t002) was positive for the sea gene, while another (ST508/CC45-t331) for the see. None of the isolates were positive for the tst gene.
4. Discussion
This is the first study on molecular characterization of S. aureus isolated from raw milk of dairy cows in Mozambique. Our findings revealed high rates of S. aureus resistant to the antibiotics commonly used in human and veterinary medicine in Manhiça District, as well as the circulation of similar genotypes causing childhood bacteremia in the study community.
The high frequency of S. aureus with similar proportions between cows from a commercial farm and the smallholder farms suggests endemicity of S. aureus among farms in Manhiça District. Comparable prevalences of S. aureus from dairy cows have been reported in other regions of Africa [33,34], Europe [35,36,37] and Asia [38,39]. The isolates showed high rates of resistance to the antibiotics commonly used in human and veterinary medicine (e.g., penicillin and tetracycline) in Manhiça District, a setting with limited access to the second line antibiotics. The high rates of resistance to penicillin and tetracycline observed among S. aureus isolated from children admitted with bacteremia in the same setting [18,40] corroborate our findings suggesting the selective pressure of resistant S. aureus due to overuse of these antibiotics in both medicines, which is not surprising. The phenotypic resistance to antibiotics other than penicillin and tetracycline was uncommon or absent among the isolates from our study, possibly due to limited availability of those antibiotics in veterinary medicine [41].
The higher resistance to penicillin and tetracycline observed among the smallholder farms compared to the commercial farm in our study could be related to the sector of production, in which the smallholder farms in Manhiça District have less access to veterinary assistance associated with the indiscriminate use of antibiotics (by farmers or other personnel who are not certified veterinarians), as those can be obtained without prescription. On the other hand, on the commercial farm veterinary assistance is frequent, although some degree of antibiotics overuse is observed. Similar to our study, high rates of resistance to penicillin and tetracycline were previously reported among S. aureus isolated from milk samples of dairy cows in Ethiopia (84–86% and 27–54%, respectively) [14,15] and in Brazil (83% and 71%, respectively) [42]. On the other hand, a recent study conducted in South Africa which analyzed S. aureus from bovine milk samples and from nasal swabs of farmers in close contact reported higher rates of resistance to penicillin (75% vs. 29%) and tetracycline (42% vs. 3%) in humans than in cattle [41].
We detected two isolates of S. aureus mecA-positive (ST508/CC45-t331 and ST1/CC1-t5388) which were cefoxitin susceptible, and three isolates tetK-positive which were tetracycline susceptible, possibly due to non-expression of the related genes, resulting in underestimation of the burden of resistant strains in the food chain [43]. The MRSA strains from our study were distinct from the well-known zoonotic livestock-associated MRSA (LA-MRSA) strains belonging to the clonal complex (CC) 398 [41]. However, the rare detection of MRSA strains from our study is in accordance with previous reports on S. aureus isolated among patients from Manhiça District and from other regions in Mozambique (1–9%) [18,19,20,40], suggesting homogeneous epidemiology of MRSA strains in humans and animals in Mozambique. Similarly, the low circulation of MDR and the absence of MRSA were reported in studies conducted in Ethiopia [14] and South Africa [16,41]. In our study, half of the isolates resistant to penicillin (52%, 13/25) were negative to detection of the blaZ gene, possibly due to a mutation of the primer-annealing site that prevented amplification [41]. The detection of other resistance determinants correlated well with the phenotypic resistance exhibited, suggesting that those are the main encoding for the antibiotic resistance observed in our context.
Data on molecular typing revealed that some strains of S. aureus can be herd-specific forming a close niche [44], be endemic or even found dispersed in a region [17]. Although there was low diversity of S. aureus observed in our study, some strains were found exclusively among the smallholder farms or in the commercial farm, while others circulated in both sectors.
We observed a high clonal structure among the isolates from our study, in which isolates from the same spa type belonged to the same ST. In addition, S. aureus isolated from distinct teats from the same animal frequently belonged to the same strain than from a distinct one, suggesting contagious transmission in the spread of the bacteria [45,46], primarily via the milking machine or milkers’ hands [13]. These sources may play an important role in cow-to-cow transmission of the same [44,47] or distinct [48] strains of S. aureus from milk or teat skin. We found two distinct strains (ST1/CC1-t5388 vs. ST152-t1299) isolated from the same milk sample, suggesting that the teats of the same animal can be colonized by unique or multiple strains, predisposing humans to infections by multiple strains of S. aureus through the ingestion of contaminated milk. In the Manhiça District, the milk commercialized by the commercial farm is pasteurized, while the one from the smallholder farms is often sold without any treatment, increasing the risk of infection for consumers. S. aureus can produce enterotoxins that are resistant to the milk treatment, predisposing humans to staphylococcal food poisoning [3] or clinical syndromes, such as toxic shock syndrome [5]. In our study, the frequency of SEs was low (3%), with only two isolates positive for sea and see genes, each. However, we only screened for genes coding for sea to see, and tst; further studies should include additional virulence factors (sef to seo, seq, ser and seu; hemolysins (e.g., hla and hlb); and exfoliative toxins (e.g., eta and etb)).
It is noteworthy that all the spa types but t355 and t5388, and all the STs from our study have previously been reported in staphylococcal bacteremia in children admitted in the Manhiça District Hospital [18,40], raising the hypothesis of potential anthroponotic transmission of S. aureus; however, additional studies are needed to verify this hypothesis. The Manhiça District is a semi-rural area where there is frequent contact between cattle and humans. Therefore, future studies in our setting should investigate the S. aureus strains isolated from cattle as well as from individuals working in close contact with these animals in order to understand the genetic relatedness of strains between hosts. In addition, our study had a limited sample size, so future studies should consider exploring additional Mozambican districts and provinces known as major dairy producers, as well as other foodborne disease-associated pathogens.
The strains from our study were previously isolated in milk samples of dairy cows in studies conducted in distinct regions across Africa (ST1, ST8, ST508 and t407) [14,16], Europe (ST1, ST5 and ST152–355) [49], Asia (ST1) [50] and in the Americas (ST1) [51]. All those STs have been isolated in dairy cows as well as in human colonization or infections [13], suggesting bacterial transmission between hosts.
In our study, the PVL gene (encoding for PVL cytotoxin) was found exclusively in the ST152 (t355 and t1299) strain, a strain that possibly originated from Africa, and then expanded throughout Europe [52]. The cytotoxin displays a specific host tropism [53], being frequently detected and associated with necrotic lesions involving the skin or mucosa in humans [29], suggesting human-to-cow transmission of S. aureus [16]. A study conducted in Ethiopia reported that all the strains belonging to t335 isolated from dairy cows were positive for the PVL gene [14].
5. Conclusions
Our findings revealed high frequency of isolation of S. aureus in raw dairy milk in the Manhiça District. The isolates showed high rates of resistance to the antibiotics commonly (penicillin and tetracycline) used in veterinary and human medicine. We observed low diversity among our isolates, in which some strains were found exclusively among the smallholder farms or in the commercial farm, as well as scattered in both. It is noteworthy that all the spa types but t355 and t5388, and all the STs from our study were reported as being associated in children admitted with bacteremia in the Manhiça District Hospital. Further investigations focusing on the molecular characterization of S. aureus isolated from cattle and farmers will provide detailed insights on the genetic relatedness between strains.
Acknowledgments
We thank the herd owners who agreed to participate in the study.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/microorganisms9081684/s1, Table S1: Primers used in this study, Table S2: Clonal structure of S. aureus isolated from raw milk in dairy cows in Manhiça District, southern Mozambique.
Author Contributions
Conceptualization, N.N., M.G., I.M. and C.T.; methodology, N.N., M.G., I.M. and C.T.; software, N.N. and M.G.; validation, H.A., I.M. and C.T.; formal analysis, N.N., M.G., H.A., I.M. and C.T.; investigation, N.N., M.G., A.M.J., A.J.M., A.C., R.V., A.O. and T.F.Z.; resources, H.A., I.M. and C.T.; data curation, H.A., I.M. and C.T.; writing—original draft preparation, N.N. and M.G.; writing—review and editing, N.N., M.G., A.M.J., A.J.M., A.C., R.V., A.O., T.F.Z., H.A., I.M. and C.T.; visualization, N.N., M.G., H.A., I.M. and C.T.; supervision, H.A., I.M. and C.T.; project administration, H.A., I.M. and C.T.; funding acquisition, H.A., I.M. and C.T. All authors have read and agreed to the published version of the manuscript.
Funding
CISM receives core funding from “Agencia Española de Cooperacion Internacional para el Desarollo (AECID)”. Marcelino Garrine was supported by the grant number 145278, from Fundação Calouste Gulbenkian “Calouste Gulbenkian Foundation”.
Institutional Review Board Statement
The study protocol was reviewed and approved by the ethics committee of the Instituto Superior de Ciências de Saúde/ISCISA (Ref: TFCMCSNNO04/18/CIBS). The ethical guidelines of ISCISA were designed in accordance with international standards.
Informed Consent Statement
A herd was recruited if the herd owner agreed to participate in the study.
Data Availability Statement
All relevant data are within the paper and its Supplementary Materials.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analysis or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Hennekinne J.-A., De Buyser M.-L., Dragacci S. Staphylococcus aureus and Its Food Poisoning Toxins: Characterization and Outbreak Investigation. FEMS Microbiol. Rev. 2012;36:815–836. doi: 10.1111/j.1574-6976.2011.00311.x. [DOI] [PubMed] [Google Scholar]
- 2.Claeys W.L., Cardoen S., Daube G., De Block J., Dewettinck K., Dierick K., De Zutter L., Huyghebaert A., Imberechts H., Thiange P., et al. Raw or Heated Cow Milk Consumption: Review of Risks and Benefits. Food Control. 2013;31:251–262. doi: 10.1016/j.foodcont.2012.09.035. [DOI] [Google Scholar]
- 3.Le Loir Y., Baron F., Gautier M. Staphylococcus aureus and Food Poisoning. Genet. Mol. Res. 2003;2:63–76. [PubMed] [Google Scholar]
- 4.Argudín M.Á., Mendoza M.C., Rodicio M.R. Food Poisoning and Staphylococcus aureus Enterotoxins. Toxins. 2010;2:1751–1773. doi: 10.3390/toxins2071751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lindsay J.A., Holden M.T.G. Staphylococcus aureus: Superbug, Super Genome? Trends Microbiol. 2004;12:378–385. doi: 10.1016/j.tim.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 6.Fitzgerald J.R. Livestock-Associated Staphylococcus aureus: Origin, Evolution and Public Health Threat. Trends Microbiol. 2012;20:192–198. doi: 10.1016/j.tim.2012.01.006. [DOI] [PubMed] [Google Scholar]
- 7.Lundberg Å., Aspán A., Nyman A., Unnerstad H., Waller K. Associations between Bacterial Genotype and Outcome of Bovine Clinical Staphylococcus aureus Mastitis. Acta Vet. Scand. 2014;56:2. doi: 10.1186/1751-0147-56-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Papadopoulos P., Papadopoulos T., Angelidis A.S., Kotzamanidis C., Zdragas A., Papa A., Filioussis G., Sergelidis D. Prevalence, Antimicrobial Susceptibility and Characterization of Staphylococcus aureus and Methicillin-Resistant Staphylococcus aureus Isolated from Dairy Industries in North-Central and North-Eastern Greece. Int. J. Food Microbiol. 2019;291:35–41. doi: 10.1016/j.ijfoodmicro.2018.11.007. [DOI] [PubMed] [Google Scholar]
- 9.Getahun K., Kelay B., Bekana M., Lobago F. Bovine Mastitis and Antibiotic Resistance Patterns in Selalle Smallholder Dairy Farms, Central Ethiopia. Trop Anim. Health Prod. 2008;40:261–268. doi: 10.1007/s11250-007-9090-5. [DOI] [PubMed] [Google Scholar]
- 10.Haftu R., Taddele H., Gugsa G., Kalayou S. Prevalence, Bacterial Causes, and Antimicrobial Susceptibility Profile of Mastitis Isolates from Cows in Large-Scale Dairy Farms of Northern Ethiopia. Trop Anim. Health Prod. 2012;44:1765–1771. doi: 10.1007/s11250-012-0135-z. [DOI] [PubMed] [Google Scholar]
- 11.Lindsay J.A., Holden M.T.G. Understanding the Rise of the Superbug: Investigation of the Evolution and Genomic Variation of Staphylococcus aureus. Funct Integr. Genom. 2006;6:186–201. doi: 10.1007/s10142-005-0019-7. [DOI] [PubMed] [Google Scholar]
- 12.Jackson C.R., Davis J.A., Barrett J.B. Prevalence and Characterization of Methicillin-Resistant Staphylococcus aureus Isolates from Retail Meat and Humans in Georgia. J. Clin. Microbiol. 2013;51:1199–1207. doi: 10.1128/JCM.03166-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zadoks R.N., Middleton J.R., McDougall S., Katholm J., Schukken Y.H. Molecular Epidemiology of Mastitis Pathogens of Dairy Cattle and Comparative Relevance to Humans. J. Mammary Gland Biol. Neoplasia. 2011;16:357–372. doi: 10.1007/s10911-011-9236-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mekonnen S.A., Lam T.J.G.M., Hoekstra J., Rutten V.P.M.G., Tessema T.S., Broens E.M., Riesebos A.E., Spaninks M.P., Koop G. Characterization of Staphylococcus aureus Isolated from Milk Samples of Dairy Cows in Small Holder Farms of North-Western Ethiopia. BMC Vet. Res. 2018;14:246. doi: 10.1186/s12917-018-1558-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eyasu T., Tesfu K., Daniel A., Haile A., Thomas S., Pamela R.F.A., Wondwossen G. Phenotypic and Genotypic Characterization of Staphylococcus aureus Isolates Recovered from Bovine Milk in Central Highlands of Ethiopia. Afr. J. Microbiol. Res. 2015;9:2209–2217. doi: 10.5897/AJMR2015.7562. [DOI] [Google Scholar]
- 16.Schmidt T., Kock M.M., Ehlers M.M. Molecular Characterization of Staphylococcus aureus Isolated from Bovine Mastitis and Close Human Contacts in South African Dairy Herds: Genetic Diversity and Inter-Species Host Transmission. Front. Microbiol. 2017;8:511. doi: 10.3389/fmicb.2017.00511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ruffing U., Alabi A., Kazimoto T., Vubil D.C., Akulenko R., Abdulla S., Alonso P., Bischoff M., Germann A., Grobusch M.P., et al. Community-Associated Staphylococcus aureus from Sub-Saharan Africa and Germany: A Cross-Sectional Geographic Correlation Study. Sci. Rep. 2017;7:154. doi: 10.1038/s41598-017-00214-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vubil D., Garrine M., Ruffing U., Acácio S., Sigaúque B., Alonso P.L., von Müller L., Herrmann M., Mandomando I. Molecular Characterization of Community Acquired Staphylococcus aureus Bacteremia in Young Children in Southern Mozambique, 2001–2009. Front. Microbiol. 2017;8:730. doi: 10.3389/fmicb.2017.00730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ceccarelli D., Mondlane J., Sale M., Salvia A.M., Folgosa E., Cappuccinelli P., Colombo M.M. Sporadic Methicillin Resistance in Community Acquired Staphylococcus aureus in Mozambique. New Microbiol. 2005;28:327–336. [PubMed] [Google Scholar]
- 20.Van der Meeren B.T., Millard P.S., Scacchetti M., Hermans M.H., Hilbink M., Concelho T.B., Ferro J.J., Wever P.C. Emergence of Methicillin Resistance and Panton-Valentine Leukocidin Positivity in Hospital- and Community-Acquired Staphylococcus aureus Infections in Beira, Mozambique. Trop. Med. Int. Health. 2014;19:169–176. doi: 10.1111/tmi.12221. [DOI] [PubMed] [Google Scholar]
- 21.Mandomando I., Espasa M., Nhampossa T., Roca A., Sigaúque B., Menéndez C., Macete E., Machevo S., Alonso P.L., Quintò L., et al. Antimicrobial Drug Resistance Trends of Bacteremia Isolates in a Rural Hospital in Southern Mozambique. Am. J. Trop. Med. Hyg. 2010;83:152–157. doi: 10.4269/ajtmh.2010.09-0578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sigaúque B., Roca A., Mandomando I., Morais L., Quintó L., Sacarlal J., Macete E., Nhamposa T., Machevo S., Aide P., et al. Community-Acquired Bacteremia among Children Admitted to a Rural Hospital in Mozambique. Pediatric Infect. Dis. J. 2009;28:108–113. doi: 10.1097/INF.0b013e318187a87d. [DOI] [PubMed] [Google Scholar]
- 23.Alonso P., Saúte F., Aponte J., Gómez-Olivé F., Nhacolo A., Thomson R., Macete E., Abacassamo F., Ventura P., Bosch X., et al. Manhiça DSS, Mozambique. In: INDEPTH, editor. Population and Health in Developing Countries Volume 1. Population, Health, and Survival at INDEPTH Sites. 1st ed. International Development Research Centre (IDRC); Ottawa, ON, Canada: 2002. pp. 189–195. [Google Scholar]
- 24.Alexopoulou K., Foka A., Petinaki E., Jelastopulu E., Dimitracopoulos G., Spiliopoulou I. Comparison of Two Commercial Methods with PCR Restriction Fragment Length Polymorphism of the Tuf Gene in the Identification of Coagulase-Negative Staphylococci. Lett. Appl. Microbiol. 2006;43:450–454. doi: 10.1111/j.1472-765X.2006.01964.x. [DOI] [PubMed] [Google Scholar]
- 25.Brakstad O.G., Aasbakk K., Maeland J.A. Detection of Staphylococcus aureus by Polymerase Chain Reaction Amplification of the Nuc Gene. J. Clin. Microbiol. 1992;30:1654–1660. doi: 10.1128/jcm.30.7.1654-1660.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.CLSI . Performance Standards for Antimicrobial Susceptibility Testing. 30th ed. Clinical and Laboratory Standards Institute; Wayne, PA, USA: 2020. Supplement M100S. [Google Scholar]
- 27.Magiorakos A.-P., Srinivasan A., Carey R.B., Carmeli Y., Falagas M.E., Giske C.G., Harbarth S., Hindler J.F., Kahlmeter G., Olsson-Liljequist B., et al. Multidrug-Resistant, Extensively Drug-Resistant and Pandrug-Resistant Bacteria: An International Expert Proposal for Interim Standard Definitions for Acquired Resistance. Clin. Microbiol. Infect. 2012;18:268–281. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- 28.Wang W., Lin X., Jiang T., Peng Z., Xu J., Yi L., Li F., Fanning S., Baloch Z. Prevalence and Characterization of Staphylococcus aureus Cultured From Raw Milk Taken From Dairy Cows With Mastitis in Beijing, China. Front. Microbiol. 2018;9:1123. doi: 10.3389/fmicb.2018.01123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lina G., Piemont Y., Godail-Gamot F., Bes M., Peter M.-O., Gauduchon V., Vandenesch F., Etienne J. Involvement of Panton-Valentine Leukocidin--Producing Staphylococcus aureus in Primary Skin Infections and Pneumonia. Clin. Infect. Dis. 1999;29:1128–1132. doi: 10.1086/313461. [DOI] [PubMed] [Google Scholar]
- 30.Aires-de-Sousa M., Boye K., de Lencastre H., Deplano A., Enright M.C., Etienne J., Friedrich A., Harmsen D., Holmes A., Huijsdens X.W., et al. High Interlaboratory Reproducibility of DNA Sequence-Based Typing of Bacteria in a Multicenter Study. J. Clin. Microbiol. 2006;44:619–621. doi: 10.1128/JCM.44.2.619-621.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Enright M.C., Day N.P., Davies C.E., Peacock S.J., Spratt B.G. Multilocus Sequence Typing for Characterization of Methicillin-Resistant and Methicillin-Susceptible Clones of Staphylococcus aureus. J. Clin. Microbiol. 2000;38:1008–1015. doi: 10.1128/JCM.38.3.1008-1015.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hunter P.R., Gaston M.A. Numerical Index of the Discriminatory Ability of Typing Systems: An Application of Simpson’s Index of Diversity. J. Clin. Microbiol. 1988;26:2465–2466. doi: 10.1128/jcm.26.11.2465-2466.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Abera M., Habte T., Aragaw K., Asmare K., Sheferaw D. Major Causes of Mastitis and Associated Risk Factors in Smallholder Dairy Farms in and around Hawassa, Southern Ethiopia. Trop Anim. Health Prod. 2012;44:1175–1179. doi: 10.1007/s11250-011-0055-3. [DOI] [PubMed] [Google Scholar]
- 34.Katsande S., Matope G., Ndengu M., Pfukenyi D.M. Prevalence of Mastitis in Dairy Cows from Smallholder Farms in Zimbabwe. Onderstepoort J. Vet. Res. 2013;80:1–7. doi: 10.4102/ojvr.v80i1.523. [DOI] [PubMed] [Google Scholar]
- 35.Olde Riekerink R.G.M., Barkema H.W., Kelton D.F., Scholl D.T. Incidence Rate of Clinical Mastitis on Canadian Dairy Farms. J. Dairy Sci. 2008;91:1366–1377. doi: 10.3168/jds.2007-0757. [DOI] [PubMed] [Google Scholar]
- 36.Ericsson Unnerstad H., Lindberg A., Persson Waller K., Ekman T., Artursson K., Nilsson-Öst M., Bengtsson B. Microbial Aetiology of Acute Clinical Mastitis and Agent-Specific Risk Factors. Vet. Microbiol. 2009;137:90–97. doi: 10.1016/j.vetmic.2008.12.005. [DOI] [PubMed] [Google Scholar]
- 37.Kadlec K., Entorf M., Peters T. Occurrence and Characteristics of Livestock-Associated Methicillin-Resistant Staphylococcus aureus in Quarter Milk Samples From Dairy Cows in Germany. Front. Microbiol. 2019;10:1–6. doi: 10.3389/fmicb.2019.01295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang F.-L., Li X.-S., He B.-X., Du Y.-L., Li G.-H., Yang B.-B., Qin-Hua H. Bovine Mastitis in Subtropical Dairy Farms, 2005-2009. J. Anim. Vet. Adv. 2011;10:68–72. doi: 10.3923/javaa.2011.68.72. [DOI] [Google Scholar]
- 39.Sudhanthiramani S., Swetha C.S., Bharathy S. Prevalence of Antibiotic Resistant Staphylococcus aureus from Raw Milk Samples Collected from the Local Vendors in the Region of Tirupathi, India. Vet. World. 2015;8:478–481. doi: 10.14202/vetworld.2015.478-481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Garrine M., Costa S.S., Mandomando I., Couto I. Antimicrobial Resistance and Molecular Typing of Staphylococcus aureus Causing Bacteraemia in Children Aged Less than 5 Years in Southern Mozambique, 2001–2019. Centro de Investigação Em Saúde de Manhiça (CISM); Maputo, Mozambique: 2021. [Google Scholar]
- 41.Schmidt T., Kock M.M., Ehlers M.M. Diversity and Antimicrobial Susceptibility Profiling of Staphylococci Isolated from Bovine Mastitis Cases and Close Human Contacts. J. Dairy Sci. 2015;98:6256–6269. doi: 10.3168/jds.2015-9715. [DOI] [PubMed] [Google Scholar]
- 42.Martini C.L., Lange C.C., Brito M.A., Ribeiro J.B., Mendonça L.C., Vaz E.K. Characterisation of Penicillin and Tetracycline Resistance in Staphylococcus aureus Isolated from Bovine Milk Samples in Minas Gerais, Brazil. J. Dairy Res. 2017;84:202–205. doi: 10.1017/S0022029917000061. [DOI] [PubMed] [Google Scholar]
- 43.Oniciuc E.-A., Nicolau A.I., Hernández M., Rodríguez-Lázaro D. Presence of Methicillin-Resistant Staphylococcus aureus in the Food Chain. Trends Food Sci. Technol. 2017;61:49–59. doi: 10.1016/j.tifs.2016.12.002. [DOI] [Google Scholar]
- 44.Capurro A., Aspán A., Ericsson Unnerstad H., Persson Waller K., Artursson K. Identification of Potential Sources of Staphylococcus aureus in Herds with Mastitis Problems. J. Dairy Sci. 2010;93:180–191. doi: 10.3168/jds.2009-2471. [DOI] [PubMed] [Google Scholar]
- 45.Sommerhäuser J., Kloppert B., Wolter W., Zschöck M., Sobiraj A., Failing K. The Epidemiology of Staphylococcus aureus Infections from Subclinical Mastitis in Dairy Cows during a Control Programme. Vet. Microbiol. 2003;96:91–102. doi: 10.1016/S0378-1135(03)00204-9. [DOI] [PubMed] [Google Scholar]
- 46.Gurjar A., Gioia G., Schukken Y., Welcome F., Zadoks R., Moroni P. Molecular Diagnostics Applied to Mastitis Problems on Dairy Farms. Vet. Clin. North. Am. Food Anim. Pract. 2012;28:565–576. doi: 10.1016/j.cvfa.2012.07.011. [DOI] [PubMed] [Google Scholar]
- 47.Haveri M., Hovinen M., Roslöf A., Pyörälä S. Molecular Types and Genetic Profiles of Staphylococcus aureus Strains Isolated from Bovine Intramammary Infections and Extramammary Sites. J. Clin. Microbiol. 2008;46:3728–3735. doi: 10.1128/JCM.00769-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zadoks R.N., van Leeuwen W.B., Kreft D., Fox L.K., Barkema H.W., Schukken Y.H., van Belkum A. Comparison of Staphylococcus aureus Isolates from Bovine and Human Skin, Milking Equipment, and Bovine Milk by Phage Typing, Pulsed-Field Gel Electrophoresis, and Binary Typing. J. Clin. Microbiol. 2002;40:3894–3902. doi: 10.1128/JCM.40.11.3894-3902.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Basanisi M.G., La Bella G., Nobili G., Franconieri I., La Salandra G. Genotyping of Methicillin-Resistant Staphylococcus aureus (MRSA) Isolated from Milk and Dairy Products in South Italy. Food Microbiol. 2017;62:141–146. doi: 10.1016/j.fm.2016.10.020. [DOI] [PubMed] [Google Scholar]
- 50.Dan M., Yehui W., Qingling M., Jun Q., Xingxing Z., Shuai M., Kuojun C., Jinsheng Z., Zibing C., Zaichao Z., et al. Antimicrobial Resistance, Virulence Gene Profile and Molecular Typing of Staphylococcus aureus Isolates from Dairy Cows in Xinjiang Province, Northwest China. J. Glob. Antimicrob. Resist. 2019;16:98–104. doi: 10.1016/j.jgar.2018.08.024. [DOI] [PubMed] [Google Scholar]
- 51.de Alves M.F.N.F., Penna B., Pereira R.F.A., Geraldo R.B., Folly E., Castro H.C., Aguiar-Alves F. First Report of Meticillin-Resistant Staphylococcus aureus Harboring MecC Gene in Milk Samples from Cows with Mastitis in Southeastern Brazil. Braz. J. Microbiol. 2020;51:2175–2179. doi: 10.1007/s42770-020-00385-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ruimy R., Maiga A., Armand-Lefevre L., Maiga I., Diallo A., Koumaré A.K., Ouattara K., Soumaré S., Gaillard K., Lucet J.-C., et al. The Carriage Population of Staphylococcus aureus from Mali Is Composed of a Combination of Pandemic Clones and the Divergent Panton-Valentine Leukocidin-Positive Genotype ST152. J. Bacteriol. (JB) 2008;190:3962–3968. doi: 10.1128/JB.01947-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Koop G., Vrieling M., Storisteanu D.M.L., Lok L.S.C., Monie T., van Wigcheren G., Raisen C., Ba X., Gleadall N., Hadjirin N., et al. Identification of LukPQ, a Novel, Equid-Adapted Leukocidin of Staphylococcus aureus. Sci. Rep. 2017;7:40660. doi: 10.1038/srep40660. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All relevant data are within the paper and its Supplementary Materials.