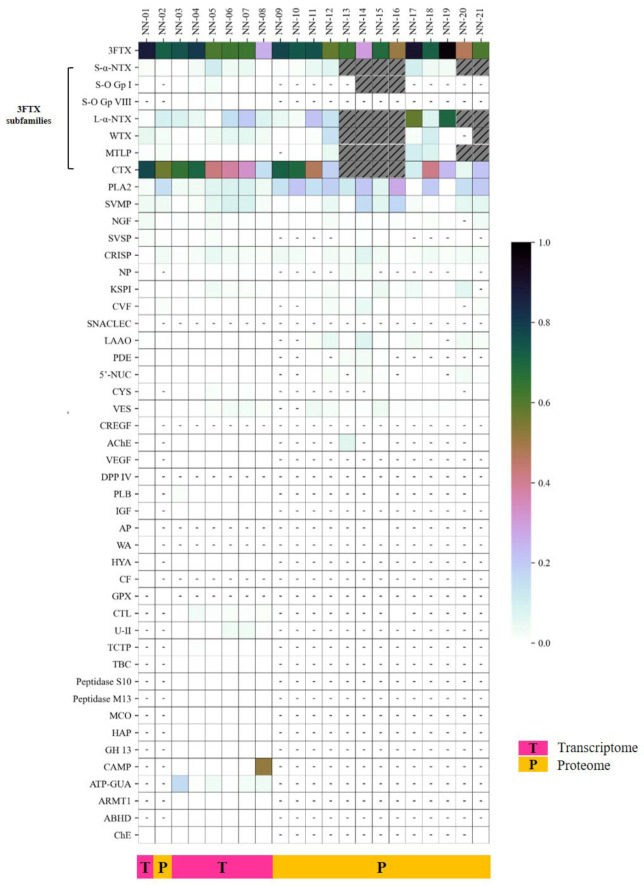
Figure 6.
Heatmap of the comparative expression profiles of toxin families derived from the venom-gland transcriptome and venom proteome of N. naja from different geographical locales. The quantitative abundances were normalized on a scale of 0–1.0 for comparison purposes across different specimens, where 1.0 depicts 100% of relative abundance, as reported in respective studies. Unreported/undetected genes or proteins were denoted in dashed lines and unsorted subtypes (mainly 3FTX) were indicated with grey stripes. Details are available in Supplementary File S3. NN-01 and NN-02 represent the relative abundance ratio of toxin families expressed in the transcriptome and proteome of NN-SL (Colombo, Sri Lanka) in the current study, respectively. NN-03 and NN-04 correspond to the relative abundance ratio of toxin families expressed in the venom gland of N. naja specimens from Kerala, India, while NN-05 to NN-08 refers to the samples from the Kentucky Reptile Zoo [31]. NN-09 and NN-10 correspond to the relative abundance ratio of N. naja venom proteins from Sri Lanka and India, respectively [7]. NN-11 corresponds to the relative abundance ratio of N. naja venom proteins from Pakistan (Sindh) [8]. NN-12 corresponds to the relative abundance ratio of N. naja venom proteins from Southern Punjab, Pakistan [44]. NN-13 corresponds to the relative abundance ratio of N. naja venom proteins from West Bengal, India [26]. NN-14 corresponds to the relative abundance ratio of N. naja venom proteins from Tamil Nadu, India [40]. NN-15 and NN-16 correspond to the relative abundance ratio of N. naja venom proteins from Western Maharashtra [93]. NN-17, NN-18 and NN-19 correspond to the relative abundance ratio of N. naja venom proteins from Indian Punjab, Rajasthan and West Bengal [6]. NN-20 corresponds to the relative abundance ratio of N. naja venom proteins from Maharashtra, India [94]. NN-21 corresponds to the relative abundance ratio of N. naja venom proteins from West Bengal, India [30]. Abbreviations: 3FTX, three-finger toxins; S-α-NTX, short chain α-neurotoxin; S-O Gp I, short chain orphan group I, S-O Gp VIII, short chain orphan group VIII; L-α-LNTX, long chain α-neurotoxin; WTX, weak neurotoxin; MTLP, muscarinic toxin-like protein; CTX, cytotoxin/cardiotoxin; PLA2, phospholipase A2; SVMP, snake venom metalloproteinases; NGF, nerve growth factor; SVSP, snake venom serine protease; CRISP, cysteine-rich secretory protein; NP, natriuretic peptide; KSPI, Kunitz-type serine protease inhibitor; CVF, cobra venom factor; SNACLEC, snake C-type lectins; LAAO, L-amino acid oxidase; PDE, phosphodiesterase; 5’-NUC, 5’-nucleotidase; CYS, cystatin; VES, vespryn; CREGF, cysteine-rich with EGF domain-like; AChE, acetylcholinesterase; VEGF, vascular endothelial growth factor; DPP IV, dipeptidylpeptidase IV; PLB, phospholipase B; IGF, insulin-like growth factor; AP, aminopeptidase; WA, waprin; HYA, hyaluronidase; CF, coagulation factor; GPX, glutathione peroxidase; CTL, C-type lectin; U-II, urotensin-II; TCTP, translationally-controlled tumor protein; TBC, Type-B carboxylesterase; MCO, multicopper oxidase; HAP, histidine acid phosphatase; GH 13, glycosyl hydrolase 13; CAMP, cathelicidin antimicrobial peptide; ATP-GUA, ATP: guanido phosphotransferase; ARMT1, ARM-transferase; ABHD, AB hydrolase; ChE, cholinesterase.