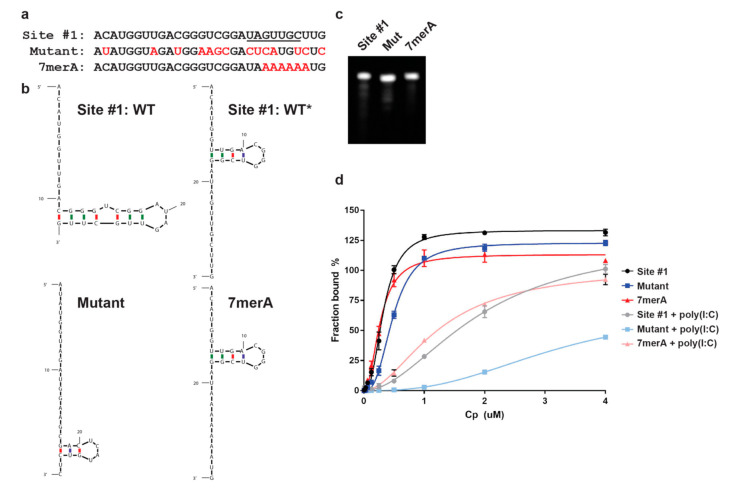
Figure 2.
SFV Cp specifically binds site #1 and 7merA RNAs. (a) Sequence alignment of site #1, Mutant, and 7merA RNAs. Red nucleotides indicate mutated residues. Underlined residues are identical in Cp’s second best gRNA binding site [20]. (b) mFold secondary structure predictions of site #1 (WT, WT*), Mutant, and 7merA RNAs. The predicted stem loop structures in WT* and 7merA are the same. WT, WT*, Mutant, and 7merA ∆G = −0.70, −0.70, −2.50, and −0.70 kcal/mol, respectively. mFold predictions were performed as described previously [20]. (c) 50 ng of site #1, Mutant, and 7merA RNAs analyzed by Urea-PAGE and SYBR Gold. (d) SFV Cp binding to Alexa488-labeled site #1, Mutant, and 7merA RNAs, −/+ poly(I:C). Data points represent the average from n = 3 independent experiments. Error bars represent the standard deviation. Data points are fit with nonlinear regression specific binding with Hill slope in Prism.