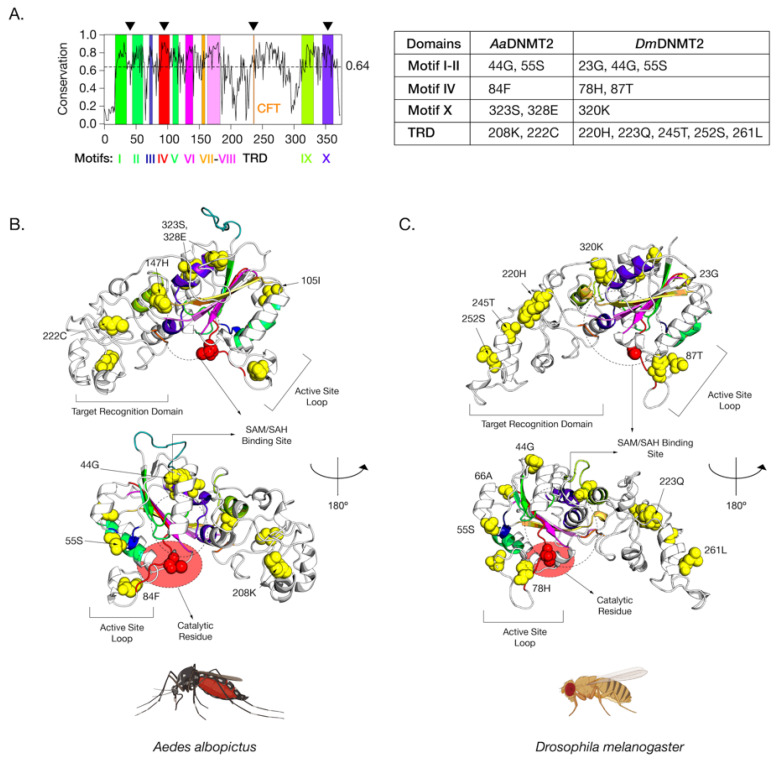
Figure 4.
Amino acid positions in Drosophila melanogaster DNMT2 potentially under positive selection. (A) Shannon conservation plot representing the degree of conservation (Y-axis) of DNMT2 orthologs present at every amino acid position (X-axis) across within DNMT2 orthologs from mosquitoes and fruit flies. Colored boxes represent known DNMT2 functional motifs and domains involved in catalytic activity and target recognition (CFT). The mean conservation score (64%) across all amino acid positions is represented by the horizontal dotted line. Black arrows present on the top represent four major regions containing most amino acid positions with evidence of positive selection and high posterior probability values (>95%). These individual amino acids are also represented in the accompanying table to the right. (B,C) Spatial distribution of sites unique to each family is represented as yellow spheres on ribbon models of (B) Aedes albopictus (9 sites) and (C) Drosophila melanogaster (12 sites) DNMT2 structures visualized in PyMOL 2.4 (Schrödinger, LLC). The catalytically active cysteine residue (Cys, C) is represented in red. The predicted substrate, i.e., S-adenosyl methionine (SAM) or S-adenosyl homocysteine (SAH), the binding region is shown as a dashed oval. Functionally important active site loop and target recognition domains are also indicated on each structure. The lower structures are rotated 180° relative to the upper ones around the vertical axis.