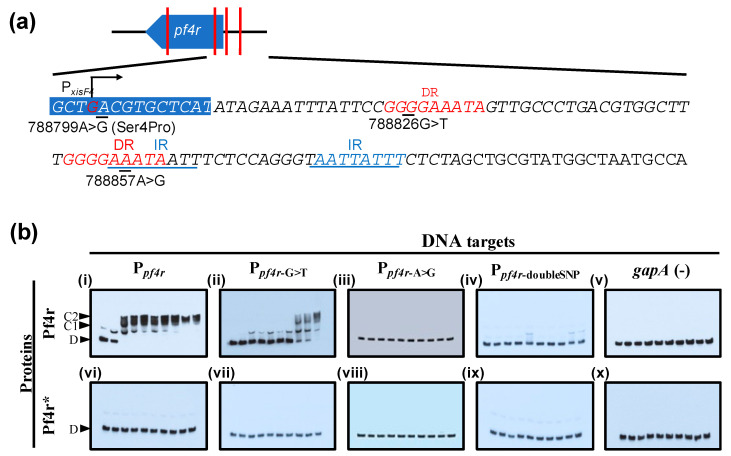
Figure 5.
EMSA of Pf4r and Pf4r* protein gradients on target DNA. (a) The positions of pf4r relative to the Pf4 genes and the single nucleotide polymorphisms within the pf4r region relative to the PAO1 genome are indicated (red vertical lines). Italicised nucleotide sequences are part of the probe used in the EMSA. The locations of the direct repeats (DR), inverted repeats (IR) and promoter start sites were as seen in Li et al. [21]. (b) Ppf4r indicates the wild-type promoter region. The Ppf4r-A>G, Ppf4r-G>T and Ppf4r-doubleSNP probes have mutations in either 788857A>G or 788826G>T, or both positions, respectively. The binding of Pf4r and Pf4r* to the selected target regions was tested using an increasing protein gradient (lanes 1-10 were 0, 10, 20, 30, 40, 50, 60, 70, 80 and 90 nM, respectively). The non-target gapA (100 bp) gene was randomly chosen as the negative control for the assays. The notations indicate free DNA (D), protein–DNA complex (C1) and protein dimer–DNA complex (C2). Pf4r restores LasB activity to the Pf4 knockout and increases frequency of variants.