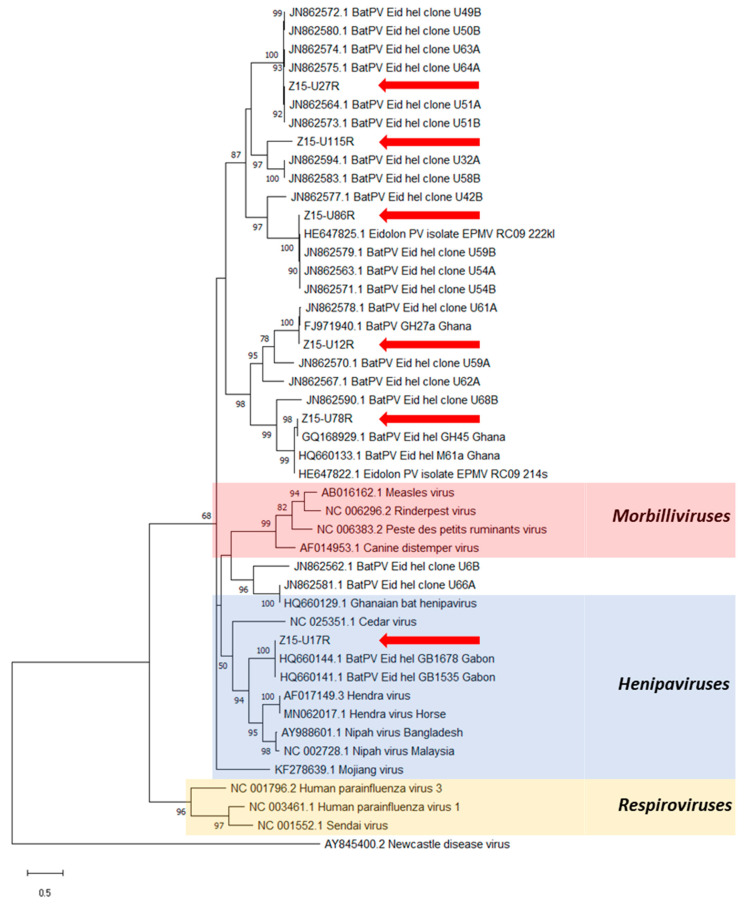
Figure 4.
Phylogenetic analysis of partial L gene sequences obtained after RMH-PCR on E. helvum urine samples (red arrows). Maximum likelihood tree with bootstrapping (1000 iterations) generated in MEGA X, using 439 bp alignment against publicly available paramyxovirus sequences (NCBI Genbank) and outgroup Newcastle disease virus. Bootstrap values for 1000 replicates are indicated as percentages (where >50%) and the number of nucleotide substitutions per site is to scale as indicated by the scale bar.