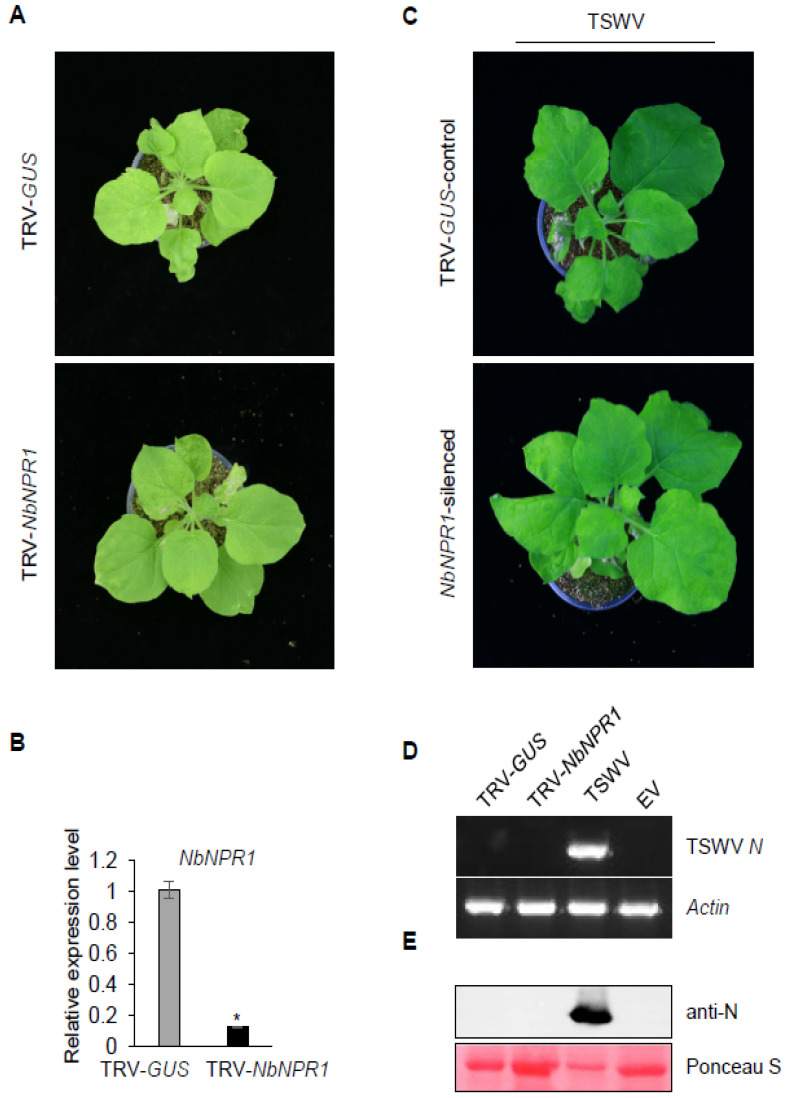
Figure 4.
Analysis of the requirement for NPR1 in Sw-5b-mediated resistance to TSWV. (A) TRV-NbNPR1- and TRV-GUS-treated Sw-5b transgenic N. benthamiana plants at 3 weeks post TRV agroinfiltration. (B) qRT-PCR analysis of the expression of NbNPR1 mRNA in newly emerged leaves of Sw-5b transgenic N. benthamiana plant treated with NPR1-silenced transgenic plants. The NbActin and NbEF1a genes were used as internal controls. Student’s t-test was used for statistical analysis (* p < 0.05). (C) Analysis of systemic infection of TSWV in Sw-5b-transgenic N. benthamiana plants silenced for NbNPR1. TRV-GUS plants were used as a control. The gene-silenced newly emerged leaves of Sw-5b-transgenic N. benthamiana plants were inoculated with TSWV. The photographs of treated plants were taken at 14 days post TSWV inoculation. (D) RT–PCR detection of TSWV N RNA in systemic leaves of Sw-5b-transgenic N. benthamiana plants silenced for NbNPR1 at 14 days post TSWV inoculation. A sample from TSWV-infected N. benthamiana tissues was used as a positive control. The NbActin gene was used as an internal reference gene. (E) Western blot analysis of TSWV N protein accumulation in the systemic leaves of Sw-5b-transgenic N. benthamiana plants silenced for NbNPR1 in panel C using specific antibodies against TSWV N. A plant sample expressed with p2300 empty vector (Vec.) was used as a negative control. Ponceau S staining was used as a protein loading control.