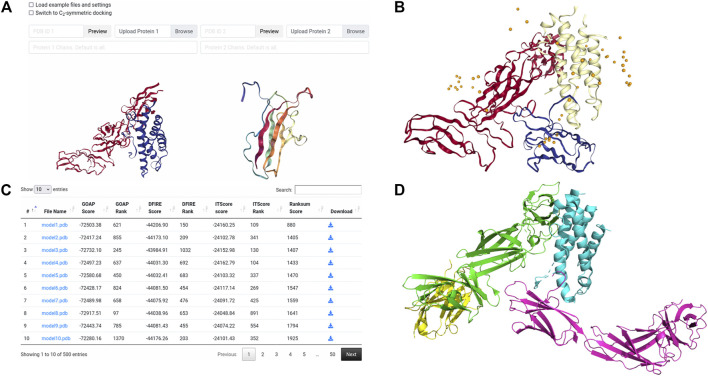
FIGURE 3.
Input and results for unconstrained docking of IL23-IL23R. (A) the input for unconstrained LZerD. The model of IL23 was uploaded as the receptor on the left, while the model of IL23R was uploaded as the ligand on the right. The chain ID selection fields are blank since we want to use all the chains. This docking run was done without constraints, so the entire constraints section is empty. (B) the results of unconstrained docking. IL23 is shown in red, while IL23R is shown in blue. The cartoon structure shown is the top-10 model, which has an of 0.28, an I-RMSD of 4.4 Å, and an L-RMSD of 7.8 Å, which is of acceptable CAPRI quality. The distribution of the top 50 ligand centroids is indicated by the orange spheres. (C) The table of model scoring information for this docking run. The ranksum score, which is used to finally rank the models, is on the right. (D) the native structure of this complex, PDB 5MZV. The non-interacting domains of IL23R and a nanobody bound to IL23 are included in the view.