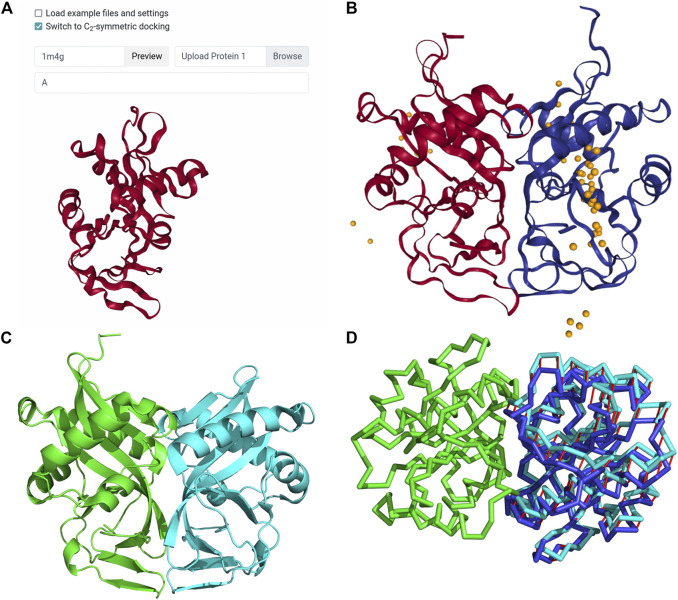
FIGURE 4.
Input and results for C2 symmetrical docking of bacterial AAC (2′). (A) The input for symmetrical LZerD. The subunit was uploaded by specifying the PDB ID 1M4G in the input field to fetch the structure from the PDB. A single chain is extracted from this structure by specifying “A” in the chain ID selection field. (B) The results of C2 symmetrical docking. The receptor and ligand are shown in red and blue respectively in the top-1 model conformation and are of course structurally identical. This model has an of 0.88, an I-RMSD of 0.95 Å, and an L-RMSD of 1.9 Å. The distribution of the top 50 ligand centroids is indicated by the orange spheres. All output models satisfy the 5.0 Å symmetricalness criterion. (C) the native structure of this complex, both chains of PDB 1M4G. Note that some N-terminal residues of chain B (cyan) are not resolved relative to chain A (green). In fact, considering all common atoms between the two native chains, they differ by 0.8 Å RMSD. (D) Visualization of the symmetricalness criterion’s satisfaction in the top-1 model viewed along the C2 symmetry axis. The subunits are shown here in Cα trace representation. Green: the receptor; cyan: the correct ligand conformation; blue: the top-1 ligand conformation; red: the deviations of the ligand from the correct conformation. This model satisfies the symmetricalness criterion with an RMSD of 2.2 Å.