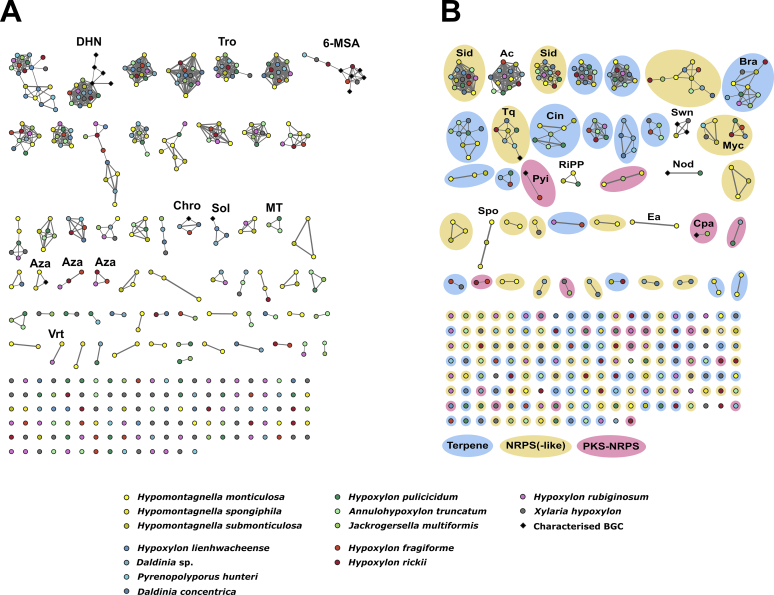
Fig. 4.
Gene cluster family (GCF) network of the 768 identified biosynthetic gene cluster (BGC) from 13 Hypoxylaceae genomes and Xylaria hypoxylon calculated by the BiG-SCAPE pipeline (cutoff value 0.4) and visualised with Cytoscape. PKS GCFs are shown on the left (A) and non-PKS GCFs are shown on the right (B). Each dot represents an identified BGC (including unclustered core genes). Characterised reference BGCs were included in the analysis (black rhombs). All BGC pairs with a distance equal or lower than the cutoff value are connected with links (thickness proportional to closeness). The layout of the figure was created according to Cytoscape's default layout algorithm ('Prefuse Force Directed Layout'), with the largest subnetworks at the top, and 'singletons', potentially unique BGCs not linked to any other BGC, shown at the bottom. The order of the singletons is random. Colors refer to species with closely related species having similar color codes. Known or predicted GCFs are labelled (6-MSA: 6-methylsalicyclic acid, Ac: alkyl citrate, Aza: azaphilone, Bra: brasilane, Chro: chromane, Cin: 1,8-cineole, Cpa: curvupallide, DHN: dihydroxynaphthalene, Ea: ergot alkaloid, MT: meroterpenoid, Myc: mycosporine-like, Nod: nodulisporic acid, Pyi: pyrichalasine, RiPP: ribosomally synthesised peptides, Sid: siderophore, Sol: solanapyrone, Spo: sporothriolide, Swn: swainsonine, Tq: terrequinone, Tro: tropolone).