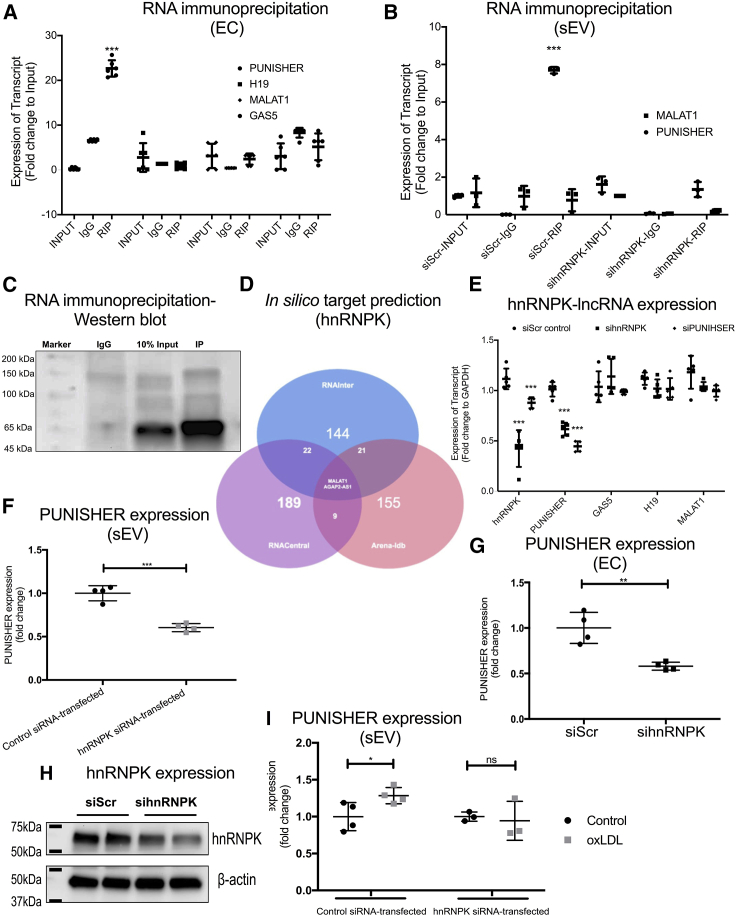
Figure 3.
Interaction between PUNISHER and heterogeneous nuclear ribonucleoprotein K (hnRNPK) mediates PUNISHER packaging into sEVs
(A) Cross-linked RNA immunoprecipitation (RIP) experiments followed by qRT-PCR quantification was performed to confirm the reciprocal binding of PUNISHER, MALAT1, H19, and GAS5 to hnRNPK in human ECs. From the RIP experiments, strong binding of hnRNPK was observed in comparison to IgG (negative control). Quantification was performed by normalizing the values with the inputs (10%). Data represent the mean ± SD (∗∗∗p < 0.001, n ≥ 4, by Student’s t test). (B) RIP experiments in sEV lysates followed by qRT-PCR quantification were performed to confirm the binding of PUNISHER, MALAT1, H19, and GAS5 in sEVs. Data represent the mean ± SD (∗∗∗p < 0.001, n ≥ 4, by Student’s t test). (C) Immunoprecipitation (IP) of hnRNPK followed by western blotting confirmed the reciprocal binding of hnRNPK to PUNISHER. The respective IgG antibody from the same species was used as an IP negative control. The enrichment was confirmed with input (10%) from the cellular lysates. (D) In silico target prediction of hnRNPK by using publicly available tools, based on the coverage score, by using RNAInter,57 RNACentral,58 and Arena-Idb59 databases. Based on their interaction scores, only MALAT1 and AGAP2-AS1 (PUNISHER) are predicted to bind to hnRNPK. (E) Expression levels of several lncRNAs (PUNIHSER, MALAT1, H19, and GAS5) upon silencing of PUNISHER/hnRNPK in ECs by using siRNAs (∗∗∗p < 0.001, n = 4, by Student’s t test). (F and G) Expression of PUNISHER upon silencing of hnRNPK in ECs and their corresponding sEVs by using siRNA (∗∗∗p < 0.001, n = 4, by Student’s t test). (H) Western blot analysis of hnRNPK protein expression was performed on the hnRNPK siRNA- or control siRNA-transfected ECs. β-actin was used as a marker. hnRNPK protein levels were assessed using ImageJ image analysis software (∗∗p < 0.01, n = 4, by Student’s t test). (I) Prior to oxLDL treatment, ECs were transfected with hnRNPK siRNA or control siRNA. PUNISHER expression was analyzed in the corresponding sEVs by qRT-PCR. GAPDH was used as an endogenous control (∗p < 0.05, n = 3−4, by Student’s t test).