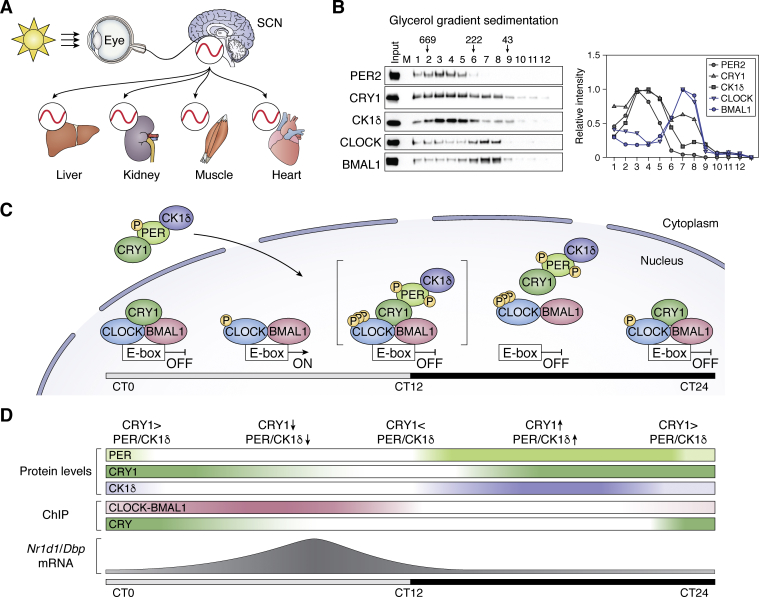
Figure 1.
Molecular mechanism of the mammalian circadian clock.A, model for circadian entrainment by light. The “master” clock in the suprachiasmatic nucleus (SCN) in the brain is entrained by neural input from photoreceptors in the retina. The master clock in turn maintains a coherent rhythmicity among clocks in peripheral tissue cells via neural signals and humoral factors. B, the positive (CLOCK-BMAL1) and negative (CRY-PER-CK1δ) arms of the TTFL are in two separate complexes. Mouse liver nuclei were harvested at ZT19 and the extract was separated by glycerol gradient velocity sedimentation along with reference proteins (thyroglobulin [669 kDa, 19S], β-amylase [222 kDa, 8.9S], and ovalbumin [43 kDa, 3.6S]). Fractions were probed by western blotting using appropriate antibodies. Left panel, western blot; right panel, quantitative scan of the western blot. CLOCK-BMAL1 sediments as a heterodimer (Mr ∼200 kDa), and PER2-CRY1- CK1δ sediments as a larger complex of Mr ∼500 kDa. C, TTFL model for the mammalian clock. The CLOCK–BMAL1 transcriptional activator binds to E-boxes at subjective dawn. At this time CRY1 is abundant and binds to the CLOCK-BMAL1-E-box complex and inhibits transcription (“Blocking type repression”). During the daytime, CRYs are degraded and CLOCK-BMAL1 activates transcription of target genes including Cry and Per. When CRY and PER accumulate, they enter the nucleus in the form of a CRY-PER-CK1δ complex, which transiently interacts with CLOCK-BMAL1-E-box (illustrated by brackets), phosphorylates CLOCK, and causes dissociation of the activator heterodimer (“Displacement type repression”). D, clock protein levels in mouse liver over the course of a circadian cycle. The levels are illustrated in the form of qualitative heatmaps, and the consequence of this clock protein change on clock-controlled Nr1d1 and Dbp gene transcription over the course of the day is plotted. Adapted with permission from Cao et al. (25).