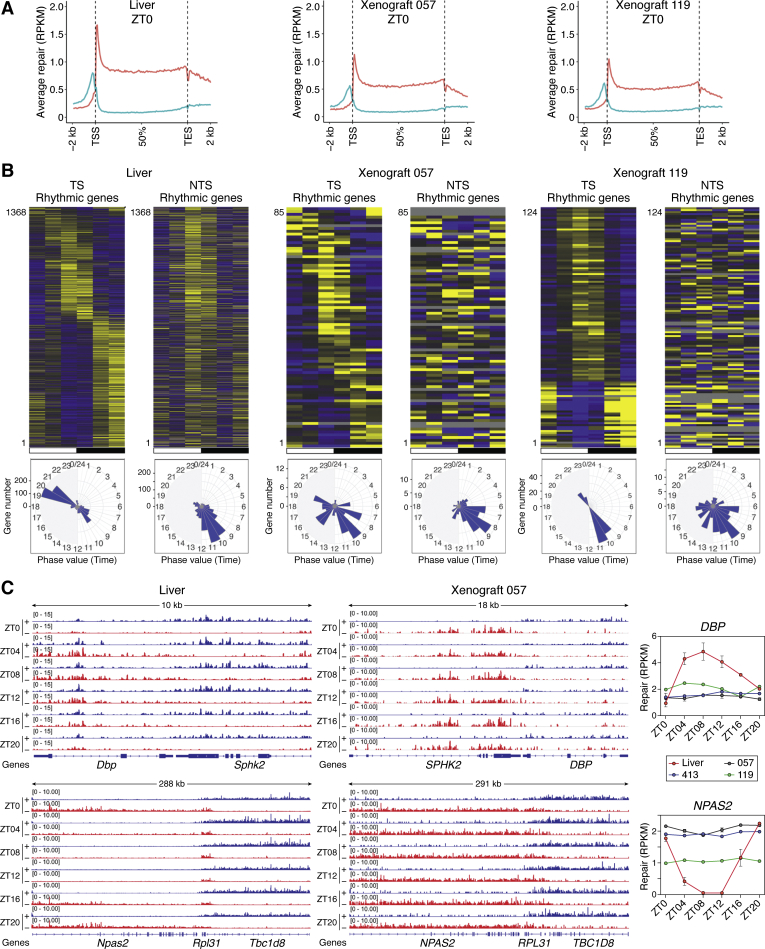
Figure 10.
Genome-wide analysis of TS and NTS repair in host liver and human colorectal cancer xenografts.A, plots of average TS and NTS repair across all genes in mouse liver and in cisplatin sensitive (057) or resistant (119) xenografts. XR-seq data obtained at ZT0 are plotted as RPKM average repair reads (y axis) along the length of a “unit gene” (x axis). The unit gene was constructed using all nonoverlapping human or mouse genes >5 kpb with a distance >5 kbp between adjacent genes. The unit gene is 100 bins in length, and values for average repair were obtained by dividing each gene into 100 bins and averaging the repair values for each successive bin for all genes from 1 to 100. Average repair 2 kbp upstream and downstream was similarly obtained. B, Heatmaps (above) and radial diagram representations (below) of circadian TS and NTS repair cycles in host liver and in cisplatin sensitive (057) and resistant (119) xenografts. In the heatmaps, each horizontal line (1368 lines for liver) represents repair of one gene from ZT0 to ZT20 at six time points. Exp/Med is, for each gene, RPKM at a given ZT time point divided by the median ZT RPKM value. The criteria for selecting the significant cyclical genes both in TS and NTS is meta2d_pvalue<0.05, meta2d_rAMP>0.1. Based on this scale, 1368, 85, and 124 genes were cyclical in host livers, xenograft 057, and xenograft 119, respectively. In an additional cisplatin resistant xenograft, 413 (not shown), 48 genes were cyclical. The host liver radial diagram (left) indicates two peaks of repair in the TS, predawn and predusk, and the NTS radial diagram to the right exhibits a single peak at ZT8-10 corresponding to our previous data (see Fig. 7). In xenografts, the TS and NTS repairs are less coherent but tend to exhibit a single peak at ZT8-11, likely due to the peak of global repair activity as described in Figure 8. C, repair of two representative circadian-controlled genes, Dbp and Npas2. The screenshots illustrate repair in liver (left) and in the cisplatin sensitive xenograft 057 (middle). It can be seen that in the liver, repair of the circadian-controlled genes (Dbp, Npas2) follows their transcriptional oscillation, while the respective neighboring genes, Sphk, Rpl31, and Tbc1d8, show constant repair over the entire circadian cycle. The graphs to the right illustrate quantitative values for TS repair as a function of circadian time for the liver, and for one sensitive (057) and two resistant (119, 413) xenografts. In contrast to the liver, rhythmic repair of the circadian-controlled genes is absent in the xenografts. From Sancar (105).