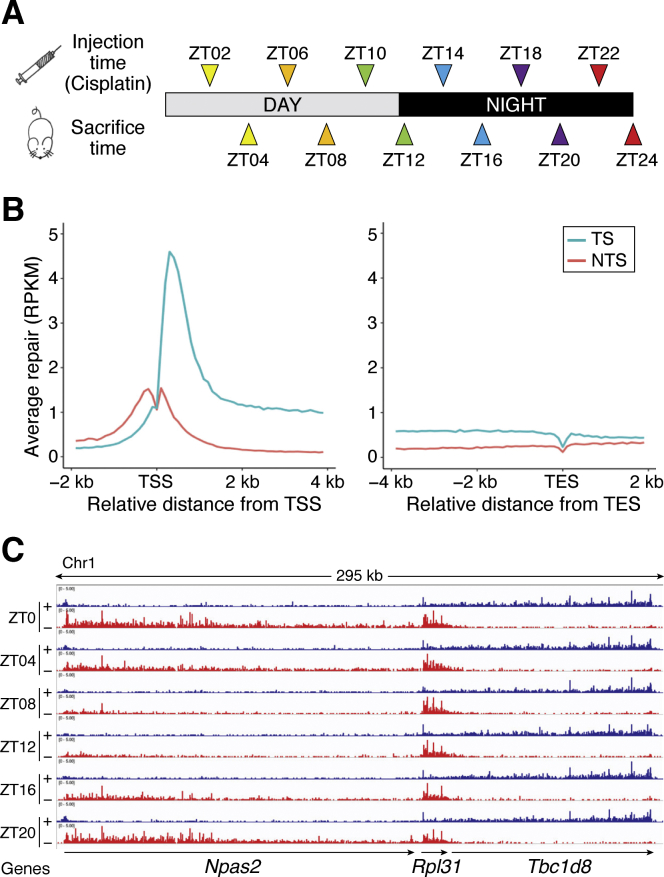
Figure 7.
Transcriptional and circadian control of excision repair of cisplatin-DNA adducts in mice.A, schematic of circadian repair experiment. Mice kept under 12-h light:12-h dark (LD 12:12) conditions were administered cisplatin at the indicated time points, and tissues were harvested 2 h later; the excision products were isolated from the liver and kidney and analyzed by XR-seq. ZT indicates circadian time where ZT0 is light-on and ZT12 is light-off. For each time point three mice were killed for XR-seq. B, genome-wide analysis of TS and NTS repair shows strong preference for TS repair in promoter-proximal regions, throughout gene bodies, and into the transcriptional end site (TES). Preferential repair reversal upstream of the transcription start sites (TSS) is due to bidirectional promoters for most mammalian genes such that the NTS in the gene body becomes the TS upstream of the TSS. The y axis shows reads per kilobase pair per million total reads (RPKM) for 100-nt windows. C, illustration showing the effect of transcription and the combined effects of the circadian clock and transcription on cisplatin repair. Repair patterns of a 295-kb region of chromosome 1 encompassing the Npas2 clock gene and two neighboring genes are shown. Blue, plus strand XR-seq repair reads; red, minus strand XR-seq repair reads. The Npas2 gene is itself clock-regulated and repair of its TS peaks at ZT20-ZT0 and troughs at ZT08. The clock output gene Rpl31 exhibits much weaker rhythmicity in repair that is delayed compared with Npas2 (peak ZT0-ZT08, minimum ZT16). Tbc1d8 exhibits high amplitude and constant TS repair over the entire course of the circadian cycle. Adapted with permission from Yang et al. (103).