FIGURE 2.
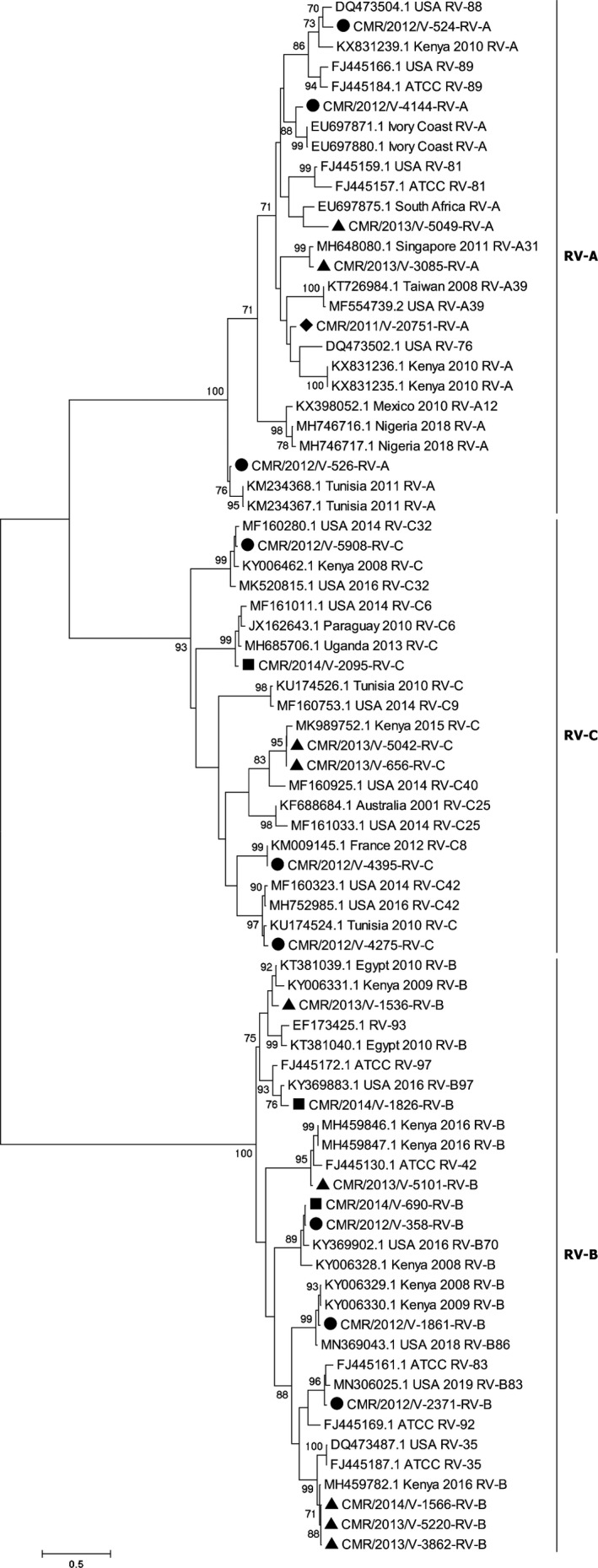
Phylogenetic tree of Rhinoviruses strains detected in Yaoundé, Cameroon, from 2011 to 2014. Multiple sequence alignment was performed with Clustal W. The rooted tree was generated based on VP4/VP2 gene nucleotide sequences using the maximum likelihood method and the best model evolution (General Time Reversible) with a Gamma distribution and assuming that a certain fraction of sites was invariable on an evolutionary level in Mega 6. The scale bars represent the frequency of nucleotide substitutions, and the numbers on the nodes of the branches are determined values for resampling bootstrap after 1000 iterations. Only values ≥70% are presented. The reference sequences of different continents, obtained on GenBank, are identified from the left to the right with the access number, the origin country and the species attributed by the author. The strains of the current study are designated from left to right by CMR (Cameroon), the year of detection and the laboratory number. Cameroon sequences are shown with (♦), (●), (▲) and (■) for the 2011, 2012, 2013 and 2014 sequences, respectively
