Abstract
Chronic hepatitis B can lead to liver cirrhosis and primary hepatocellular carcinoma. The present study aimed to investigate whether C-X-C motif chemokine receptor 3 (CXCR3) regulates the genes in Toll-like receptors (TLRs)/myeloid differentiation primary response protein 88 (MyD88) signaling pathway in the development of hepatitis B into cirrhosis and liver cancer in vitro. A hepatitis B virus (HBV) overexpression lentivirus was constructed and infected into a LX-2 cell line to obtain stable HBV-overexpressing cells (named HBV-LX-2 cells). The CXCR3 gene was knocked down using small interfering RNA in HBV-LX-2 cells. Cell Counting Kit-8 assays, cell scratch tests and flow cytometry were used to detect cell proliferation, migration and apoptosis, respectively. The levels of IL-1β and IL-6 in serum samples of patients with liver cancer were measured via ELISA, and the collagen content in liver cancer tissues was detected using Masson staining. Western blotting was used to detect the expression levels of proteins in the TLRs/MyD88 signaling pathway. Excessive fibrosis was identified in the liver cancer tissues, and the serum levels of IL-6 and IL-1β were abnormally increased in patients with liver cancer. It was found that interfering with CXCR3 inhibited cell proliferation and migration, as well as promoted the apoptosis of HBV-LX-2 cells. Moreover, interfering with CXCR3 inhibited the expression levels of collagen type I α 1 chain and the proteins in the TLRs/MyD88 pathway. In conclusion, CXCR3 knockdown could inhibit the expression levels of proteins in the TLR4/MyD88 signaling pathway, decrease cell proliferation and migration, and promote cell apoptosis, thus inhibiting the development of liver cirrhosis to liver cancer.
Keywords: C-X-C motif chemokine receptor 3, hepatitis B virus, Toll-like receptors, myeloid differentiation primary response protein 88, liver cirrhosis
Introduction
Chronic hepatitis B virus (HBV) infection is the main cause of different degrees of hepatic inflammation. Moreover, chronic hepatitis can lead to the development of liver cirrhosis and hepatocellular carcinoma (HCC) (1,2). According to a report from the World Health Organization, ~257 million individuals are chronically infected with HBV worldwide, and 0.74 million individuals die of HBV-related liver cirrhosis annually (2).
Chemokines and their receptors serve essential roles in the development of HCC (3). C-X-C motif chemokine receptor 3 (CXCR3) belongs to the G-protein coupled receptor 1 family, and it is expressed in a variety of cells, such as T1 helper T cells, CD8 T cells, natural killer (NK) T cells, NK cells, dendritic cells and some cancer cells, such as colon and breast cancer cells (4,5). CXCR3 specifically binds with C-X-C motif chemokine ligand 9 (CXCL9) to promote the migration and invasion of cancer cells (6). In addition, it activates the PI3K/Akt signaling pathway to regulate cell proliferation, apoptosis and angiogenesis (7,8). Furthermore, CXCR3 is rapidly increased after interferon stimulation, and it promotes the migration of CD4+ T and CD8+ T cells to the inflamed tissues (9,10). It has been shown that the expression of CXCR3 on T cells mainly exerts anti-tumor effects, while its expression on tumor cells mainly promotes cancer metastasis (11,12).
HBV causes hepatitis and HCC mainly via the host's immune and inflammatory response (13). Innate immunity and adaptive immunity serve important roles in HBV infection (14). Toll-like receptors (TLRs) are innate immune recognition receptors that can resist infection by recognizing microbial cell wall components and triggering immune response (15,16). Myeloid differentiation primary response protein 88 (MyD88) is a cytosolic adapter protein that is involved in the TLR signaling pathway (17). TLR4 can activate the TIR domain containing adaptor protein/MyD88 signal transduction pathway, and induce the production of proinflammatory cytokines, such as IL-6, IL-10, IL-12 and TNF-α, via NF-κB and activator protein 1 (18). In hepatocytes, the TLR/MyD88 signaling pathway is activated upon contact with HBV (19,20). Moreover, the enhanced production of IL-6 and TNF-α could inhibit HBV replication and prevent the death of HBV-infected hepatocytes (21,22). Host recognition of HBV also leads to the activation of NLR family pyrin domain containing 3 (NALP3), and subsequently to the release of IL-1β. Abnormal secretion of IL-1β can aggravate the inflammatory reaction, and mature IL-1β binds to its receptor, IL-1βR, and reactivates MyD88 (23,24).
Previous studies have mainly focused on the role of CXCR3 or TLRs in cancer, and have reported their respective roles in the development of hepatitis B into HCC (25–27). However, the regulatory relationship between CXCR3 and the TLRs/MyD88 signaling pathway has been rarely reported. Thus, the present study aimed to investigate whether CXCR3 regulated the genes in the TLRs/MyD88 signaling pathway, and to examine the effect of CXCR3 on the proliferation, migration and apoptosis of hepatic stellate cells infected with HBV. The current study may reveal the regulatory relationship between CXCR3 and the TLRs/MyD88 signaling pathway in the carcinogenesis of liver cirrhosis.
Materials and methods
Materials
The human hepatic stellate cell line LX-2 (cat. no. BNCC341586) was obtained from BeNa Culture Collection (Neijing Beina Chunglian Institute of Biotechnology). Rabbit anti-CXCR3 (cat. no. ab71864), rabbit anti-TLR4 (cat. no. ab13556), rabbit anti-MyD88 (cat. no. ab133739), rabbit anti-NALP3 (cat. no. ab214185), rabbit anti-COL1A1 (cat. no. ab34710), rabbit anti-baculoviral IAP repeat containing 5 (BIRC5; cat. no. ab469) and rabbit anti-GAPDH (cat. no. ab181602) antibodies were purchased from Abcam. The goat anti-rabbit IgG (H+L) (HRP) antibody (cat. no. ZB-2301) was from Beijing Zhongshan Goldenbridge Biotechnology Co., Ltd. FBS (cat. no. 10099141) and Opti-MEM™ (cat. no. 31985-062) were purchased from Gibco (Thermo Fisher Scientific, Inc.). RIPA buffer (cat. no. 1053) was from Applygen Technologies, Inc. DMEM (cat. no. KGM12800S) was purchased from Nanjing KeyGen Biotech Co., Ltd. pCDH-CMV-MCS-EF1-copGFP-T2A-Puro vector (cat. no. P0268) was provided by Wuhan Miaoling Biotechnology Co., Ltd. SuperSignal® West Pico Chemiluminescent Substrate (cat. no. 34077) was from Thermo Fisher Scientific, Inc.
Masson's Trichrome stain kit (cat. no. G1340) and BSA (cat. no. A8020) were purchased from Beijing Solarbio Science & Technology Co., Ltd. IL-6 (cat. no. m1058097-1) and IL-1β (cat. no. m105859-1) ELISA kits were from Shanghai Enzyme-linked Biotechnology Co., Ltd. The Annexin V-APC/7-AAD apoptosis kit (cat. no. AP105-100-kit) was from Multisciences (Lianke) Biotech Co., Ltd. Ultrapure RNA kit (cat. no. CW0581M), HiFiScript cDNA Synthesis kit (cat. no. CW2569M), UltraSYBR Mixture (cat. no. CW0957M) and BCA Protein Assay kit (cat. no. CW0014S) were purchased from CoW in Biosciences. The Cell Counting Kit (CCK)-8 (cat. no. KGA317) was from Nanjing KeyGen Biotech Co., Ltd.
Sample collection
The study was approved by The Ethics Committee of Hwa Mei Hospital, University of Chinese Academy of Sciences (Ningbo, China; approval no. PJ-NBEY-KY-2018-016-01), and written informed consent was obtained from all participants. In total, 20 subjects were recruited into this study, including 10 healthy controls, 4 patients with liver cancer and 6 patients with liver cancer with cirrhosis. The clinical and pathological features of the patients with cancer were summarized in Table I. Liver cancer tissues and the paracancerous tissues were collected from 6 patients with liver cancer who had a history of liver cirrhosis. A total of 10 ml serum samples were collected from 10 patients with liver cancer and 10 healthy controls. The controls were healthy subjects who received physical examinations. The patients with cancer were recruited at Hwa Mei Hospital between January 2019 and May 2019.
Table I.
Clinical and pathological features of the patients with cancer.
| No. | Sex | Age, years | Clinical diagnosis | Histological type | TNM stage | Metastasis |
|---|---|---|---|---|---|---|
| 1 | Male | 63 | Malignant tumor of liver | Moderately differentiated HCC | I | No |
| 2 | Male | 58 | Liver tumor, HB | Moderately differentiated HCC | II | No |
| 3 | Male | 62 | Liver tumor, active CHB | Poorly differentiated HCC | II | No |
| 4 | Female | 77 | Liver tumor | Moderately differentiated HCC | II | No |
| 5 | Male | 69 | Malignant tumor of liver | Well-moderately differentiated HCC | II | No |
| 6 | Male | 56 | Malignant tumor of liver | Moderately-poorly differentiated HCC | II | No |
| 7 | Female | 58 | Liver tumor | Well-moderately differentiated HCC | I | No |
| 8 | Male | 60 | Liver tumor | Moderately differentiated HCC | III | Yes (blood) |
| 9 | Male | 53 | Liver tumor | Poorly differentiated HCC | II | No |
| 10 | Female | 77 | Liver tumor | Moderately-poorly differentiated HCC | II | No |
HCC, hepatocellular carcinoma; CHB, chronic hepatitis B; HB, hepatitis B.
The inclusion criteria were as follows: Subjects join the study voluntarily and sign informed consent; aged 18–75 years; any sex; patients with liver cancer diagnosed by histopathology/cytology or clinical diagnosis; ≥1 lesion that could be measured according to the Response Evaluation Criteria In Solid Tumors v1.1 (28); liver function status as Child-Pugh Class A or B (score ≤7); Eastern Cooperative Oncology Group physical status score of 0–1; life expectancy ≥12 weeks; laboratory examination findings of: i) Blood system function, absolute neutrophil count ≥1.5×109/l, platelet count ≥75×109/l, hemoglobin ≥90 g/l; ii) liver function, total bilirubin ≤1.5× the upper limit of normal value (ULN), serum albumin ≥28 g/l, alanine aminotransferase and aspartate transaminase ≤5× ULN; and iii) renal function, serum creatinine ≤1.5× ULN, endogenous creatinine clearance rate ≥60 ml/min; normal blood pressure or use of one antihypertensive drug can that can make the blood pressure ≤150/90 mmHg; and the patients are willing and able to follow up. The exclusion criteria were as follows: Brain metastasis; tumor thrombus in the main portal vein or inferior vena cava; presence of double or multiple cancer types; moderate or severe ascites; history of bleeding tendency or thrombosis; history of severe cardiovascular disease; history of unhealed wound or ulcer, or fracture within 3 months; laboratory abnormalities: i) Hyponatremia (sodium <130 mmol/l); baseline serum potassium <3.5 mmol/l; ii) abnormal thyroid function, and thyroid function that cannot be maintained in the normal range with drugs; iii) HIV positive; pregnant or lactating women; and patients considered unsuitable for the trial by the researchers.
Generation of stable cell line
The human hepatic stellate cell line LX-2 was cultured in DMEM supplemented with 10% FBS. The cells were maintained at 37°C in the presence of 5% CO2. The sequences of the HBV (gene ID, 944566) gene were synthesized by General Biosystems (Anhui) Co., Ltd., and inserted into a lentiviral expression vector pCDH-CMV-MCS-EF1-copGFP-T2A-Puro. LX-2 cells were transfected with HBV lentivirus vector using 8 µg/ml polyamine at 37°C. Cells were observed under a fluorescence microscope (MF53; Guangzhou Mingmei Photoelectric Technology Co., Ltd.), and the images were taken under white and green fluorescent light. At 72 h following transfection, cells were incubated with 5 µg/ml puromycin at 37°C for 72 h, followed by selection with 2 µg/ml puromycin. The puromycin resistant clones were tested for HBV expression via PCR and western blotting. The positive stable clones were maintained in the presence of puromycin.
Transient transfection
The sequences of all small interfering (si)RNAs used in this study were: CXCR3-siRNA-F1 forward, 5′-GCUAAAUGACGCCGAGGUUTT-3′ and reverse, 5′-AACCUCGGCGUCAUUUAGCTT-3′; CXCR3-siRNA-F2 forward, 5′-AGAGAGGGCUCCAGAGGCATT-3′ and reverse, 5′-UGCCUCUGGAGCCCUCUCUTT-3′; CXCR3-siRNA-F3 forward, 5′-CUGGAGAACUUCAGCUCUUTT-3′ and reverse, 5′-AAGAGCUGAAGUUCUCCAGTT-3′; and siRNA-negative control (NC) forward, 5′-UUCUCCGAACGUGUCACGUTT-3′ and reverse, 5′-ACGUGACACGUUCGGAGAATT-3′. For transient CXCR3 knockdown, LX-2 cells with stable HBV overexpression were transiently transfected with CXCR3 siRNAs using Lipofectamine® 3000 (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's instructions. In brief, cell culture medium was replaced with serum-free medium when cells were cultured to 70% confluence. Then, 5 µl Lipofectamine 3000 and 12.5 µl siRNA were diluted in 125 µl Opti-MEM, and then incubated at room temperature for 5 min. The diluted Lipofectamine was mixed with the diluted siRNA and incubated at room temperature for 15 min. The complexes were added to the culture medium. After 6 h, the transfection medium was replaced with culture medium. Subsequent experiments were performed 48 h following transfection.
Reverse transcription (RT)-PCR and RT-quantitative (q)PCR
Total RNA from the LX-2 cells was extracted with the Ultrapure RNA kit according to the manufacturer's instructions. RT reactions were performed with the HiFiScript cDNA Synthesis kit in a total reaction volume of 20 µl containing 50 pg-5 µg RNA template, 4 µl dNTP mix, 4 µl 5X RT buffer, 2 µl DTT, 2 µl primer mix and 1 µl HiFiScript. The reaction was incubated at 42°C for 15 min, 85°C for 5 min and finally held at 4°C. The primers for HBV were 5′-TCTCAGCAATGTCAACGACC-3′ (forward) and 5′-AATTTATGCCTACAGCCTCCT-3′ (reverse). The cDNA of HBV was subjected to PCR, and the RT-PCR products were analyzed by electrophoresis on a 2% agarose gel. The primers used were as follows: CXCR3 forward, 5′-AATGACGCCGAGGTTGC-3′ and reverse, 5′-CCAGAGCCAAAGACCCACT-3′; and GAPDH forward, 5′-TGACTTCAACAGCGACACCCA-3′ and reverse, 5′-CACCCTGTTGCTGTAGCCAAA-3′. qPCR was performed using the UltraSYBR Mixture on a CFX Connect™ Real-time PCR Detection system (Bio-Rad Laboratories, Inc.), under the following thermocycling conditions: Initial denaturation at 95°C for 10 min, followed by 40 cycles of 95°C for 10 sec, 58°C for 30 sec and 72°C for 30 sec, and final extension at 72°C for 10 min. All samples were run in duplicate, and the mRNA expression level of CXCR3 was analyzed using the 2−ΔΔCq method (29).
Western blot analysis
Protein in cells was extracted with ice-cold RIPA buffer. The samples were then centrifuged at 13,780 × g at 4°C for 10 min, and the supernatant was transferred to a new tube. The protein concentration was detected with a BCA Protein assay kit. A total of 40 µg protein per lane was loaded and subsequently separated by 10% SDS-PAGE and transferred to PVDF membranes. Membranes were blocked in 3% non-fat milk at 37°C for 1 h before incubation with the primary antibodies at 4°C overnight, including rabbit anti-CXCR3 (1:1,000), rabbit anti-TLR4 (1:1,000), rabbit anti-MyD88 (1:1,000), rabbit anti-NALP3 (1:500), rabbit anti-COL1A1 (1:1,000), rabbit anti-BIRC5 (1:5,000) and rabbit anti-GAPDH (1:10,000). After washing with 1X TBS-Tween-20 (0.1%) three times, each for 10 min, the membranes were incubated with the goat anti-rabbit IgG (H+L) (HRP) secondary antibody (1:2,000) at 37°C for 2 h. Protein bands were detected using the SuperSignal® West Pico Chemiluminescent Substrate and analyzed via Image-Pro Plus software (version 6.0; Media Cybernetics, Inc.).
CCK-8 assay
The CCK-8 kit was used according to the manufacturer's instructions. Following transfection, 10 µl CCK-8 solution was added to the 96-well plates and incubated for 2 h at 37°C. The optical density (OD) value of each well was detected at 48 and 72 h at 450 nm wavelength using an enzyme-labeled instrument, and the cell viability was calculated.
Cell apoptosis assay
In total, 1–3×106 cells were collected and centrifuged with 1 ml PBS twice at 210 × g for 3 min at room temperature. Then, 5X Binding Buffer was diluted to 1X Binding Buffer with double distilled water, and the cells were then resuspended in 1X Binding Buffer. Subsequently, 3 µl annexin V-APC and 5 µl 7-AAD were added to incubate the cells in the dark at room temperature for 10 min. Then, 200 µl pre-cold 1X Binding Buffer was added into each tube and mixed well. Cell apoptotic rates were calculated by detecting the percentage of early and late apoptotic cells using a flow cytometer (NovoCyte® 2060R; ACEA Bioscience, Inc.) with NovoExpress software (version 1.3.4; Agilent Technologies, Inc.).
Cell migration assay
At the start of the assay, cells on either side of the would were at 95–100% confluence. A sterile 10-µl pipette tip was used to make a single scratch, then the culture medium was discarded and the HBV-LX-2 cells were washed three times with PBS. Serum-free medium was added to each well, and the scratch of each well was imaged at 0, 24 and 48 h under an inverted fluorescence microscope (MF53; Guangzhou Mingmei Photoelectric Technology Co., Ltd.). The cell migration rate was calculated according to the width of the scratch using Image-Pro Plus software (version 5.1.0.20; Media Cybernetics, Inc.).
ELISA
IL-6 and IL-1β levels in serum were assessed using the ELISA kits according to the manufacturer's instructions. Briefly, 50 µl Standard Substance with different concentrations at 48, 24, 12, 6, 3 and 0 µg/ml were added into the standard wells. The samples were incubated with 100 µl enzyme labeled reagent at 37°C for 60 min. Following washing with PBS, 50 µl Developer A and 50 µl Developer B were added into each well, shaken and mixed gently, and then the color was developed at 37°C for 15 min. Subsequently, 50 µl termination solution was added to stop the reaction. The OD of each well was measured at 450 nm on an automatic microplate reader (WD-2102B; Beijing Liuyi Biotechnology Co., Ltd.).
Masson staining
Tissues were fixed in 10% formaldehyde at 4°C for 48 h. Liver sections (4 µm) were deparaffinized, rehydrated, stained in Weigert's iron hematoxylin solution at room temperature for 10 min and then differentiated with acid ethanol for 5–15 sec. After washing in running water for 10 min, the slides were stained in Masson blue solution for at 37°C for 5 min and rinsed in distilled water, which was followed by staining in ponceau-acid fuchsin solution at 37°C for 8 min and differentiation in phosphomolybdic acid solution for 2 min. The slides were then transferred directly to aniline blue solution and stained at 37°C for 2 min, washed with weak acid working solution for 1 min and quickly dehydrated three times in 95% ethanol and anhydrous ethanol. Finally, the slides were sealed and observed under an optical microscope (magnification, ×200 and ×400).
Statistical analysis
All statistical analyses were performed using SPSS 19.0 software (IBM Corp.). Data are presented as the mean ± standard deviation of three independent experiments. One-way ANOVA was used to compare the differences between multiple groups, followed by the post hoc Student-Newman-Keuls test and Tukey's test. P<0.05 was considered to indicate a statistically significant difference.
Results
Establishment of LX-2 cell line stably overexpressing HBV
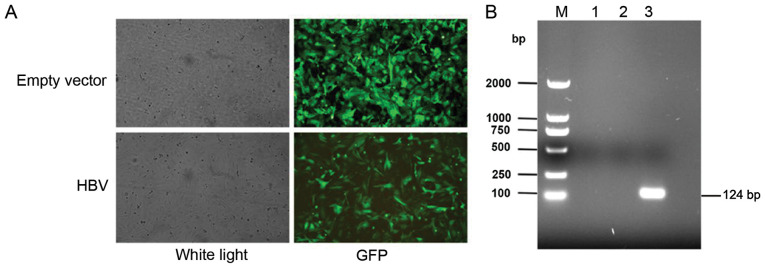
The HBV lentivirus vector was transfected into LX-2 cells, and the cells were observed using a fluorescence microscope. Representative images of the green fluorescent protein fluorescence from LX-2 cells following transfection of the empty lentiviral vector and the HBV lentivirus vector are shown in Fig. 1A. The HBV-overexpressing stable cell line was selected by puromycin, and the RT-PCR results demonstrated that HBV mRNA expression was notably increased in the HBV-transfected cells compared with the empty lentiviral vector-transfected cells and the control cells (Fig. 1B).
Figure 1.
Establishment of the HBV-overexpressing stable cell line. (A) GFP fluorescence from LX-2 cells following transfection of the empty lentivirus vector and the HBV lentivirus vector. Magnification, ×100. (B) The electrophoretogram of reverse transcription-PCR products of HBV. M, marker; lane 1, Control; lane 2, empty vector; lane 3, HBV. GFP, green fluorescent protein; HBV, hepatitis B virus.
Interference of CXCR3 gene expression
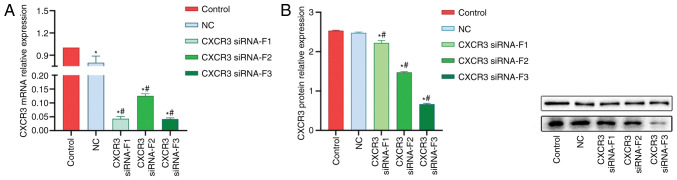
A total of three CXCR3 siRNAs were synthesized and transfected into LX-2 cells stably expressing HBV (named HBV-LX-2 cells). The interference efficiency of siRNAs was detected via RT-qPCR and western blot analyses. The RT-qPCR results demonstrated that the interference efficiencies of all the three siRNAs were >50% (Fig. 2A). The results from western blot analysis revealed that siRNA-F3 showed the strongest interference efficiency on CXCR3 protein expression, which was >50% (Fig. 2B).
Figure 2.
Interference of CXCR3 gene expression. (A) mRNA and (B) protein expression levels of CXCR3 in LX-2 cells stably expressing HBV (named HBV-LX-2 cells) following transfection with siRNAs. *P<0.05 vs. Control; #P<0.05 vs. NC. CXCR3, C-X-C motif chemokine receptor 3; HBV, hepatitis B virus; NC, siRNA-negative control; siRNA, small interfering RNA.
Interference with CXCR3 inhibits HBV-LX-2 cell viability and migration and promotes cell apoptosis
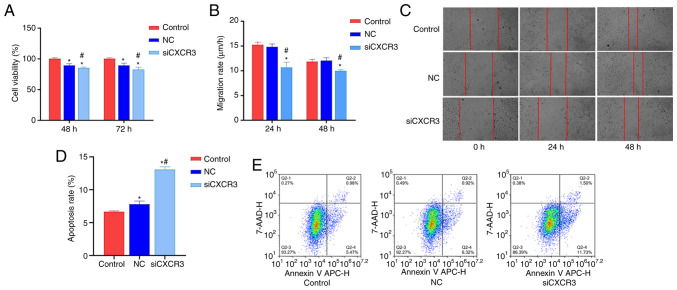
In order to examine the effect of CXCR3 on the viability, migration and apoptosis of HBV-LX-2 cells, CXCR3 siRNA-F3 was transfected into HBV-LX-2 cells. The cells were then subjected to CCK-8 assays, and cell migration and cell apoptosis analyses. Compared with the control group and NC group, the viability of the siCXCR3 group was significantly decreased at 48 and 72 h (Fig. 3A), and the cell migratory rate significantly decreased at 24 and 48 h (Fig. 3B and C). Moreover, the cell apoptotic rate was significantly increased in the siCXCR3 group compared with the control group and the NC group (Fig. 3D and E).
Figure 3.
Interference with CXCR3 inhibits HBV-LX-2 cell viability and migration and promotes cell apoptosis. (A) Interference with CXCR3 inhibits HBV-LX-2 cell viability. (B) Interference with CXCR3 inhibits the migratory rate of HBV-LX-2 cells. (C) Representative images of HBV-LX-2 cell migration following transfection with siCXCR3. Magnification, ×100. (D) Interference with CXCR3 promotes HBV-LX-2 cell apoptosis. (E) Representative images of HBV-LX-2 cell apoptosis following transfection with siCXCR3. *P<0.05 vs. Control; #P<0.05 vs. NC. CXCR3, C-X-C motif chemokine receptor 3; HBV, hepatitis B virus; siRNA/si, small interfering RNA; NC, siRNA-negative control.
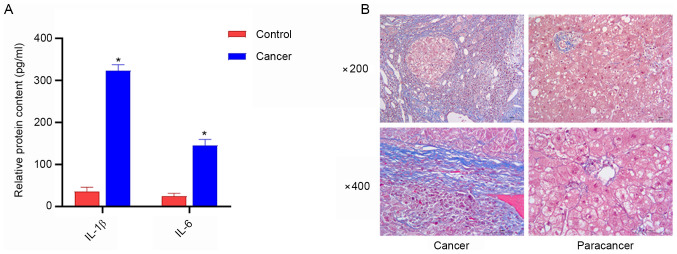
Serum levels of IL-1β and IL-6 are increased in patients with liver cancer, and a high amount of collagen fibers exists in liver cancer tissues
ELISA was used to detect serum levels of IL-β and IL-6 in patients with liver cancer, and the degree of liver fibrosis was examined using Masson staining. The results of ELISA demonstrated that the serum levels of IL-1β and IL-6 in patients with liver cancer were higher compared with those in the healthy controls (Fig. 4A). Furthermore, Masson staining revealed that liver cancer tissues contained a large number of collagen fibers, while the content of collagen fibers in paracancerous tissues was reduced (Fig. 4B).
Figure 4.
Serum levels of IL-1β and IL-6, and the degree of liver fibrosis in patients with liver cancer. (A) Serum levels of IL-1β and IL-6 were increased in patients with cancer. *P<0.05 vs. Control. (B) Masson staining of the liver cancer tissues and paracancerous tissues.
Interference with CXCR3 inhibits the expression levels of COL1A1 and the proteins in the TLRs/MyD88 pathway in HBV-LX-2 cells
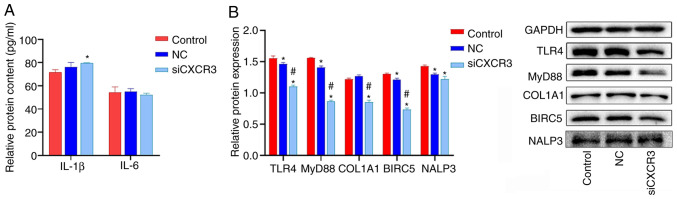
CXCR3 siRNA was transfected into LX-2 cells overexpressing HBV, and the cells were subjected to western blot analysis. The results demonstrated that the protein expression levels of TLR4, MyD88, COL1A1 and BIRC5 were significantly lower in the siCXCR3 group compared with those in the control group and the NC group (Fig. 5B). Compared with the control group, the expression level of NALP3 protein was significantly decreased in the siCXCR3 group. However, there was no significant difference in NALP3 protein expression between siCXCR3 group and NC group.
Figure 5.
Effect of siCXCR3 on the expression levels of COL1A1 and the proteins in the TLRs/MyD88 pathway in HBV-LX-2 cells. (A) Interference with CXCR3 increases the expression of IL-1β in HBV-LX-2 cells, as determined by ELISA. (B) Interference with CXCR3 inhibits the protein levels of TLR4, MyD88, COL1A1 and BIRC5 in HBV-LX-2 cells, as determined via western blotting. *P<0.05 vs. Control; #P<0.05 vs. NC. CXCR3, C-X-C motif chemokine receptor 3; siRNA/si, small interfering RNA; NC, siRNA-negative control; TLR, Toll-like receptor; MyD88, myeloid differentiation primary response protein 88; COL1A1, collagen type I α 1 chain; BIRC5, baculoviral IAP repeat containing 5; NALP3, NLR family pyrin domain containing 3.
The levels of IL-6 and IL-1β in HBV-LX-2 cells were detected via ELISA. IL-1β and IL-6 levels in LX-2 cells were lower than those in the serum of patients with liver cancer (Figs. 4A and 5A). IL-6 levels were significantly increased in the siCXCR3 group compared with the control group, but there was no significant difference in the level of IL-6 between siCXCR3 group and NC group. Compared with the control group and NC group, the level of IL-1β was not significantly changed in the siCXCR3 group (Fig. 5A).
Discussion
In the process of hepatic fibrosis, hepatic stellate cells differentiate into proliferative and contractile myofibroblasts, as well as secrete COL1A1 and other proteins that constitute pathological fibrous tissues (30). Fibrosis can lead to liver cancer, and CXCR3 serves an important role in this process. In previous studies, CXCR3 has been reported to promote the proliferation and migration of cancer cells, including gastric, colorectal and tongue squamous cell carcinoma cells (6–8). The present study established a HBV-overexpressing stable cell line using hepatic stellate cells LX-2. RNA interference-mediated knockdown of CXCR3 was performed in HBV-LX-2 cells, and the results demonstrated that CXCR3 was significantly decreased at both the mRNA and protein levels in cells following transfection with CXCR3 siRNA. Furthermore, it was found that interference with the expression level of CXCR3 could inhibit cell proliferation and migration and promote cell apoptosis. Previous studies have shown that CXCR3 was upregulated in primary and metastatic cancer cells (8,31,32). In glioma cells, CXCR3 activates PI3K/Akt via the G protein subunit to increase intracellular calcium level, thereby promoting the proliferation, migration and invasion of cancer cells (31). Moreover, CXCR3 is highly expressed in gastric cancer tissues with lymph node metastasis, suggesting that CXCR3 can promote lymph node metastasis (8). In tongue squamous cell carcinoma, CXCL9/CXCR3 promotes the invasion and proliferation of cancer cells by activating the Akt signaling pathway (32). All these reports suggest that CXCR3 can promote cell proliferation, migration and invasion. In the present study, the results demonstrated that interfering with the expression level of CXCR3 inhibited cell proliferation and migration. This finding was consistent with the role of CXCR3 in other cancer cells (8,31,32), indicating that interference with CXCR3 can inhibit the carcinogenesis of hepatic stellate cells overexpressing HBV.
Chronic HBV infection may progress to liver cirrhosis and hepatocellular carcinoma (33). In the present study, Masson staining was performed on the liver cancer tissues of liver cancer cases with HBV infection. A large number of collagen fibers were observed in the liver cancer tissues, but the amount of collagen fibers in adjacent tissues was small, indicating that the excessive accumulation of collagen fibers in liver cancer tissues may lead to liver cirrhosis. Moreover, it was found the serum levels of IL-1β and IL-6 were increased in the patients with liver cancer. These findings suggested that the liver cancer induced by hepatitis B can promote the expression of inflammatory factors. Excessive IL-1β and IL-6 will aggravate the inflammatory reaction and promote the development of liver cirrhosis (24). In the current study, LX-2 cells stably overexpressing HBV were cultured in vitro. It was found that the levels of IL-1β and IL-6 in LX-2 cells were much lower than those in the serum of patients with liver cancer. Moreover, CXCR3 inhibition did not significantly downregulate the levels of IL-1β and IL-6 in HBV-LX-2 cells.
Previous studies have reported that CXCR3 is involved in the activity of cancer cells, such as proliferation, migration and invasion (6–8). In addition, CXCR3 can induce the chemotaxis of immune cells, especially T cells (34,35). Upregulated CXCR3 can promote the migration of T cells to specific organs (9,36), induce the secretion of cytokines such as IL-6 and IFN-γ, and cause inflammation. In the current experiments, only one cell line was cultured in vitro. Therefore, CXCR3 could not exert its effect on T cell chemotaxis to induce inflammatory response. It has been reported that IL-1β is expressed not only in immune cells, but also in other cells (37). The expression and activation of IL-1β is controlled by two different signaling pathways (38,39). The first signaling pathway is NF-κB-mediated IL-1β section, and this pathway is induced by TLR ligands. Another signaling pathway is the cleavage of the precursor IL-1β to mature IL-1β via caspase-1, which is mediated by NALP3 (38,39). In the present study, it was found that TLR4 and MyD88 were significantly downregulated in cells following CXCR3 interference, but the expression level of NLRP3, as well as the levels of IL-1β and IL-6 did not change significantly. This finding suggested that in HBV-LX-2 cells, the expression of IL-1β and IL-6 was mediated by NLRP3, but not by TLR4/MyD88. Moreover, it was identified that CXCR3 could not directly regulate the expression level of NLRP3, so IL-1β and IL-6 were almost unchanged in vitro.
Although there was no significant change in inflammatory factors, COL1A1 protein expression was significantly downregulated in HBV-LX-2 cells following transfection with CXCR3 siRNA, indicating that CXCR3 knockdown could decrease the synthesis of collagen and reduce the degree of liver fibrosis. Previous studies have shown that COL1A1 expression was upregulated in gastric cancer cells and promoted cell invasion and metastasis (40,41). In cervical cancer, COL1A1 can inhibit the apoptosis of cancer cells via the PI3K pathway, while interference with COL1A1 can significantly promote the apoptosis of cancer cells (42). The present results were consistent with those of previous studies in that interference with CXCR3 downregulates COL1A1 protein expression, promotes apoptosis and inhibits the migration of HBV-LX-2 cells (8,32,40,42). Similar results were observed with regards to BIRC5 protein expression. BIRC5 is an anti-apoptotic protein, which is widely expressed in malignant tumors, such as gastric cancer, lung cancer, colon cancer and ovarian cancer (43,44). BIRC5 can affect cell division and proliferation, as well as inhibit apoptosis of cancer cells (45). Moreover, interference with BIRC5 triggers the apoptosis of cancer cells (46). The present study demonstrated that CXCR3 siRNA could downregulate BIRC5 protein expression, inhibit cell proliferation and promote cell apoptosis, suggesting that interference with CXCR3 could inhibit the differentiation of HBV-LX-2 cells to cancer cells.
The present study only detected the expression level of CXCR3 in HBV-LX-2 cells and solely investigated the role of CXCR3 at the cellular level. As a limitation of this study, protein expression studies were not performed on patient tissues. Further studies may be conducted in clinical tissues and animal model tissues to perform correlation analysis between CXCR3 and the inflammatory factors, genes in the TLR4/MyD88 signaling pathway and other factors involved in the process of liver cirrhosis to liver cancer.
In conclusion, the present study demonstrated that CXCR3 knockdown can inhibit the expression levels of TLR4/MyD88, BIRC and COL1A1, as well as inhibit cell proliferation and migration and promote cell apoptosis, so as to inhibit the development of liver cirrhosis to liver cancer. It was also found that knockdown of CXCR3 had little effect on the expression levels of NALP3 and the related inflammatory factors IL-1β and IL-6. Thus, the role of CXCR3 in regulating the inflammatory factors IL-1β and IL-6 during the process of liver cirrhosis to liver cancer should be further studied.
Acknowledgements
Not applicable.
Funding Statement
This study was funded by Medical and Health Project of Zhejiang Province (grant no. 2019KY179) and Joint Construction of Key Medical Disciplines in Zhejiang Province (grant no. 2016-7).
Funding
This study was funded by Medical and Health Project of Zhejiang Province (grant no. 2019KY179) and Joint Construction of Key Medical Disciplines in Zhejiang Province (grant no. 2016-7).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
DW and GY conceived and designed the experiments, as well as prepared the manuscript. BC, XS, YM and JL conducted the experiments. AH and YH contributed to data collection and analyzed the data. DW and GY are confirm the authenticity of all the raw data. All authors read and approved the final manuscript.
Ethics approval and consent to participate
This study was approved by the Ethics Committee of Hwa Mei Hospital, University of Chinese Academy of Sciences (Ningbo, China). All patients provided written informed consent in compliance with the code of ethics of the World Medical Association (Declaration of Helsinki).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.McMahon BJ. Chronic hepatitis B virus infection. Med Clin North Am. 2014;98:39–54. doi: 10.1016/j.mcna.2013.08.004. [DOI] [PubMed] [Google Scholar]
- 3.Liang CM, Chen L, Hu H, Ma HY, Gao LL, Qin J, Zhong CP. Chemokines and their receptors play important roles in the development of hepatocellular carcinoma. World J Hepatol. 2015;7:1390–1402. doi: 10.4254/wjh.v7.i10.1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tokunaga R, Zhang W, Naseem M, Puccini A, Berger MD, Soni S, McSkane M, Baba H, Lenz HJ. CXCL9, CXCL10, CXCL11/CXCR3 axis for immune activation-A target for novel cancer therapy. Cancer Treat Rev. 2018;63:40–47. doi: 10.1016/j.ctrv.2017.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Karin N. CXCR3 ligands in cancer and autoimmunity, chemoattraction of effector T cells, and beyond. Front Immunol. 2020;11:976. doi: 10.3389/fimmu.2020.00976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Humblin E, Kamphorst AO. CXCR3-CXCL9: It's all in the tumor. Immunity. 2019;50:1347–1349. doi: 10.1016/j.immuni.2019.05.013. [DOI] [PubMed] [Google Scholar]
- 7.Friedl P, Alexander S. Cancer invasion and the microenvironment: Plasticity and reciprocity. Cell. 2011;147:992–1009. doi: 10.1016/j.cell.2011.11.016. [DOI] [PubMed] [Google Scholar]
- 8.Zhou H, Wu J, Wang T, Zhang X, Liu D. CXCL10/CXCR3 axis promotes the invasion of gastric cancer via PI3K/AKT pathway-dependent MMPs production. Biomed Pharmacother. 2016;82:479–488. doi: 10.1016/j.biopha.2016.04.069. [DOI] [PubMed] [Google Scholar]
- 9.Kohlmeier JE, Cookenham T, Miller SC, Roberts AD, Christensen JP, Thomsen AR, Woodland DL. CXCR3 directs antigen-specific effector CD4+ T cell migration to the lung during parainfluenza virus infection. J Immunol. 2009;183:4378–4384. doi: 10.4049/jimmunol.0902022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marshall A, Celentano A, Cirillo N, McCullough M, Porter S. Tissue-specific regulation of CXCL9/10/11 chemokines in keratinocytes: Implications for oral inflammatory disease. PLoS One. 2017;12:e0172821. doi: 10.1371/journal.pone.0172821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abron JD, Singh NP, Murphy AE, Mishra MK, Price RL, Nagarkatti M, Nagarkatti PS, Singh UP. Differential role of CXCR3 in inflammation and colorectal cancer. Oncotarget. 2018;9:17928–17936. doi: 10.18632/oncotarget.24730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Y, Xu L, Peng M. CXCR3 is a prognostic marker and a potential target for patients with solid tumors: A meta-analysis. Onco Targets Ther. 2018;11:1045–1054. doi: 10.2147/OTT.S157421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Villarrubia VG, Alvarez-Mon M, Chirigos MA, Herrerías JM. Hepatitis B virus (HBV) and the inflammatory/immune response. I. The natural environment of the antigen presentation and immunologic chaos induced by the virus. Rev Esp Enferm Dig. 1997;89:919–928. (In Spanish) [PubMed] [Google Scholar]
- 14.Fisicaro P, Valdatta C, Boni C, Massari M, Mori C, Zerbini A, Orlandini A, Sacchelli L, Missale G, Ferrari C. Early kinetics of innate and adaptive immune responses during hepatitis B virus infection. Gut. 2009;58:974–982. doi: 10.1136/gut.2008.163600. [DOI] [PubMed] [Google Scholar]
- 15.Nosratabadi R, Alavian SM, Zare-Bidaki M, Shahrokhi VM, Arababadi MK. Innate immunity related pathogen recognition receptors and chronic hepatitis B infection. Mol Immunol. 2017;90:64–73. doi: 10.1016/j.molimm.2017.07.002. [DOI] [PubMed] [Google Scholar]
- 16.Gao D, Li W. Structures and recognition modes of toll-like receptors. Proteins. 2017;85:3–9. doi: 10.1002/prot.25179. [DOI] [PubMed] [Google Scholar]
- 17.Takeda K, Akira S. TLR signaling pathways. Semin Immunol. 2004;16:3–9. doi: 10.1016/j.smim.2003.10.003. [DOI] [PubMed] [Google Scholar]
- 18.Firmal P, Shah VK, Chattopadhyay S. Insight Into TLR4-mediated immunomodulation in normal pregnancy and related disorders. Front Immunol. 2020;11:807. doi: 10.3389/fimmu.2020.00807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mylona EE, Mouktaroudi M, Crisan TO, Makri S, Pistiki A, Georgitsi M, Savva A, Netea MG, van der Meer JW, Giamarellos-Bourboulis EJ, Joosten LA. Enhanced interleukin-1β production of PBMCs from patients with gout after stimulation with toll-like receptor-2 ligands and urate crystals. Arthritis Res Ther. 2012;14:R158. doi: 10.1186/ar3898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guo H, Jiang D, Ma D, Chang J, Dougherty AM, Cuconati A, Block TM, Guo JT. Activation of pattern recognition receptor-mediated innate immunity inhibits the replication of hepatitis B virus in human hepatocyte-derived cells. J Virol. 2009;83:847–858. doi: 10.1128/JVI.02008-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hösel M, Quasdorff M, Wiegmann K, Webb D, Zedler U, Broxtermann M, Tedjokusumo R, Esser K, Arzberger S, Kirschning CJ, et al. Not interferon, but interleukin-6 controls early gene expression in hepatitis B virus infection. Hepatology. 2009;50:1773–1782. doi: 10.1002/hep.23226. [DOI] [PubMed] [Google Scholar]
- 22.Xu Y, Köck J, Lu Y, Yang D, Lu M, Zhao X. Suppression of hepatitis B virus replication in Tupaia hepatocytes by tumor necrosis factor alpha of Tupaia belangeri. Comp Immunol Microbiol Infect Dis. 2011;34:361–368. doi: 10.1016/j.cimid.2011.05.003. [DOI] [PubMed] [Google Scholar]
- 23.Gupta P, Barthwal MK. IL-1 β genesis: The art of regulating the regulator. Cell Mol Immunol. 2018;15:998–1000. doi: 10.1038/s41423-018-0054-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mitroulis I, Kambas K, Ritis K. Neutrophils, IL-1β, and gout: Is there a link? Semin Immunopathol. 2013;35:501–512. doi: 10.1007/s00281-013-0361-0. [DOI] [PubMed] [Google Scholar]
- 25.Elia G, Fallahi P. Hepatocellular carcinoma and CXCR3 chemokines: A narrative review. Clin Ter. 2017;168:e37–e41. doi: 10.7417/CT.2017.1980. [DOI] [PubMed] [Google Scholar]
- 26.Xu HZ, Liu YP, Guleng B, Ren JL. Hepatitis B virus-related hepatocellular carcinoma: Pathogenic mechanisms and novel therapeutic interventions. Gastrointest Tumors. 2014;1:135–145. doi: 10.1159/000365307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zare-Bidaki M, Tsukiyama-Kohara K, Arababadi MK. Toll-like receptor 4 and hepatitis B infection: Molecular mechanisms and pathogenesis. Viral Immunol. 2014;27:321–326. doi: 10.1089/vim.2014.0039. [DOI] [PubMed] [Google Scholar]
- 28.Eisenhauer EA, Therasse P, Bogaerts J, Schwartz LH, Sargent D, Ford R, Dancey J, Arbuck S, Gwyther S, Mooney M, et al. New response evaluation criteria in solid tumours: Revised RECIST guideline (version 1.1) Eur J Cancer. 2009;45:228–247. doi: 10.1016/j.ejca.2008.10.026. [DOI] [PubMed] [Google Scholar]
- 29.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 30.Brenner DA, Waterboer T, Choi SK, Lindquist JN, Stefanovic B, Burchardt E, Yamauchi M, Gillan A, Rippe RA. New aspects of hepatic fibrosis. J Hepatol. 2000;32(Suppl 1):S32–S38. doi: 10.1016/S0168-8278(00)80413-4. [DOI] [PubMed] [Google Scholar]
- 31.Zhou YQ, Liu DQ, Chen SP, Sun J, Zhou XR, Xing C, Ye DW, Tian YK. The role of CXCR3 in neurological diseases. Curr Neuropharmacol. 2019;17:142–150. doi: 10.2174/1570159X15666171109161140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li Z, Liu J, Li L, Shao S, Wu J, Bian L, He Y. Epithelial mesenchymal transition induced by the CXCL9/CXCR3 axis through AKT activation promotes invasion and metastasis in tongue squamous cell carcinoma. Oncol Rep. 2018;39:1356–1368. doi: 10.3892/or.2017.6169. [DOI] [PubMed] [Google Scholar]
- 33.Papatheodoridis GV, Chan HL, Hansen BE, Janssen HL, Lampertico P. Risk of hepatocellular carcinoma in chronic hepatitis B: Assessment and modification with current antiviral therapy. J Hepatol. 2015;62:956–967. doi: 10.1016/j.jhep.2015.01.002. [DOI] [PubMed] [Google Scholar]
- 34.Zeng Z, Li L, Chen Y, Wei H, Sun R, Tian Z. Interferon-γ facilitates hepatic antiviral T cell retention for the maintenance of liver-induced systemic tolerance. J Exp Med. 2016;213:1079–1093. doi: 10.1084/jem.20151218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Benigni G, Dimitrova P, Antonangeli F, Sanseviero E, Milanova V, Blom A, van Lent P, Morrone S, Santoni A, Bernardin G. CXCR3/CXCL10 axis regulates neutrophil-NK cell cross-talk determining the severity of experimental osteoarthritis. J Immunol. 2017;198:2115–2124. doi: 10.4049/jimmunol.1601359. [DOI] [PubMed] [Google Scholar]
- 36.Masopust D, Vezys V, Marzo AL, Lefrançois L. Preferential localization of effector memory cells in nonlymphoid tissue. Science. 2001;291:2413–2417. doi: 10.1126/science.1058867. [DOI] [PubMed] [Google Scholar]
- 37.Rébé C, Ghiringhelli F. Interleukin-1β and cancer. Cancers (Basel) 2020;12:1791. doi: 10.3390/cancers12071791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Walsh JG, Muruve DA, Power C. Inflammasomes in the CNS. Nat Rev Neurosci. 2014;15:84–97. doi: 10.1038/nrn3638. [DOI] [PubMed] [Google Scholar]
- 39.Dinarello CA. Immunological and inflammatory functions of the interleukin-1 family. Ann Rev Immunol. 2009;27:519–550. doi: 10.1146/annurev.immunol.021908.132612. [DOI] [PubMed] [Google Scholar]
- 40.Li AQ, Si JM, Shang Y, Gan LH, Guo L, Zhou TH. Construction of COL1A1 short hairpin RNA vector and its effect on cell proliferation and migration of gastric cancer cells. Zhejiang Da Xue Xue Bao Yi Xue Ban. 2010;39:257–263. doi: 10.3785/j.issn.1008-9292.2010.03.008. (In Chinese) [DOI] [PubMed] [Google Scholar]
- 41.Sun H. Identification of key genes associated with gastric cancer based on DNA microarray data. Oncol Lett. 2016;11:525–530. doi: 10.3892/ol.2015.3929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu S, Liao G, Li G. Regulatory effects of COL1A1 on apoptosis induced by radiation in cervical cancer cells. Cancer Cell Int. 2017;17:73. doi: 10.1186/s12935-017-0443-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ambrosini G, Adida C, Altieri DC. A novel anti-apoptosis gene, survivin, expressed in cancer and lymphoma. Nat Med. 1997;3:917–921. doi: 10.1038/nm0897-917. [DOI] [PubMed] [Google Scholar]
- 44.Gunaldi M, Isiksacan N, Kocoglu H, Okuturlar Y, Gunaldi O, Topcu TO, Karabulut M. The value of serum survivin level in early diagnosis of cancer. J Cancer Res Ther. 2018;14:570–573. doi: 10.4103/0973-1482.171369. [DOI] [PubMed] [Google Scholar]
- 45.Wheatley SP, Altieri DC. Survivin at a glance. J Cell Sci. 2019;132:jcs223826. doi: 10.1242/jcs.223826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li F, Aljahdali I, Ling X. Cancer therapeutics using survivin BIRC5 as a target: What can we do after over two decades of study? J Exp Clin Cancer Res. 2019;38:368. doi: 10.1186/s13046-019-1362-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.