Figure 3. Genetics of cancer predisposition genes in pediatric acute lymphoblastic leukemia.
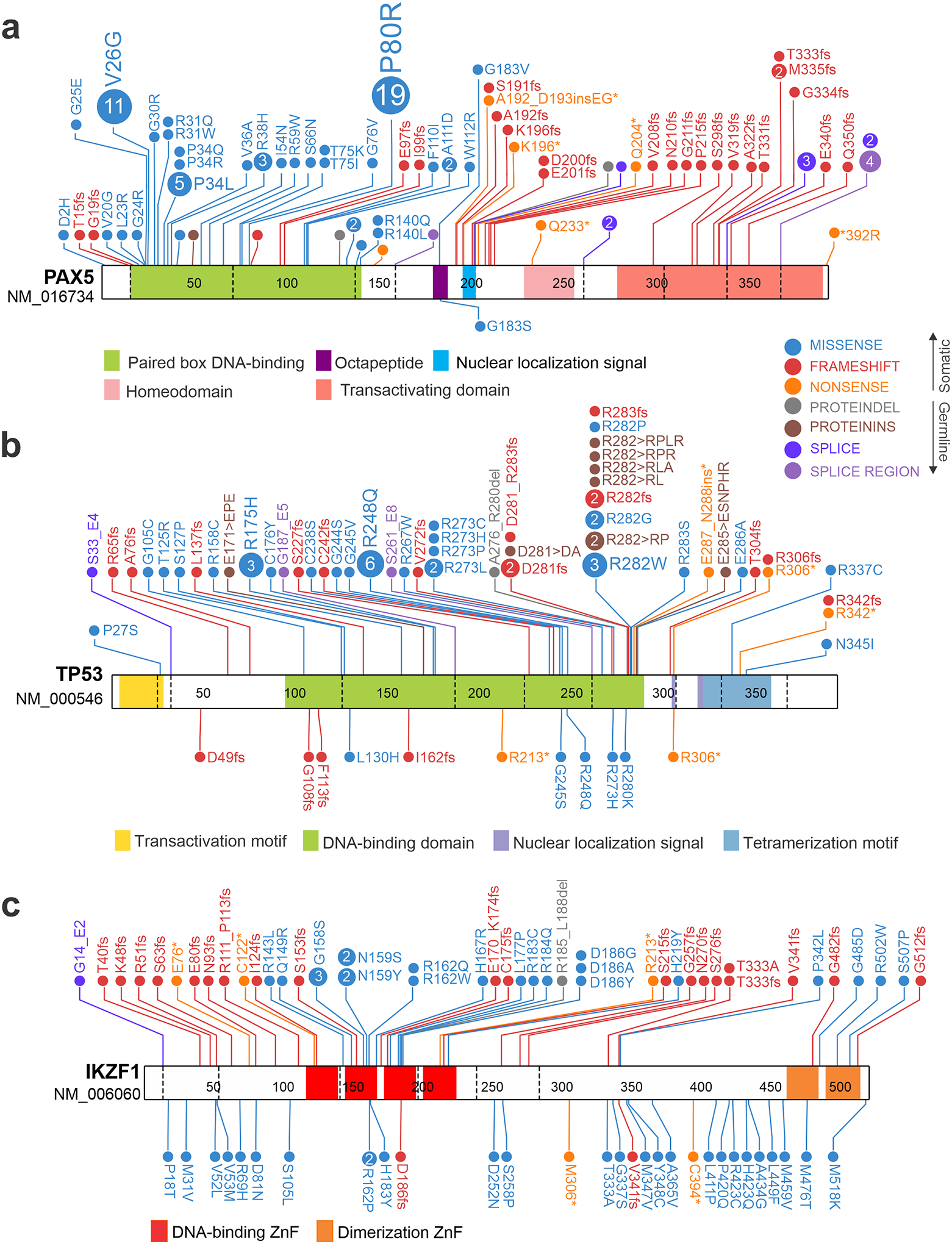
Schematics and domain architectures of proteins encoded by genes harboring germline and somatic mutations in pediatric acute lymphoblastic leukemia (ALL), including PAX5 (a), TP53 (b) and IKZF1 (c). The germline mutations are shown below the protein schematic and the somatic mutations on top. (a) PAX5 somatic variants are predominantly missense variants in the DNA binding domain and truncations that remove the transactivation domain. By contrast, a single residue has been observed mutated in familial ALL, G183S in the octapeptide domain. (b) Germline TP53 variants are shown for hypodiploid ALL, which involve the DNA binding domain and nuclear localization sequences. (c) Differences in distribution in IKZF1 variants are observed between germline and somatic variants. Somatic variants are most commonly N terminal truncating mutations, DNA binding domain missense mutations (most commonly in the second and third DNA-binding zinc fingers (ZnFs), which are required for DNA binding), and distal truncating and C-terminal ZnF missense mutation. In contrast, germline variants are predominantly located outside of ZnFs (possibly as these would not be tolerated as germline events) but in functional assays such as protein localization, adhesion and drug resistance, are commonly as deleterious or more deleterious than somatic variants. Germline data taken from refs 8,12,143; somatic mutation data updated from ref. 180.
