While DNMT3A mutations (DNMT3AMT) mostly represent early event in the clonal hierarchy, a broad analysis of the clonal architecture of DNMT3A-associated neoplasia has not been conducted across disease sub-entities and/or mutation types. Moreover, the prognostic impact of DNMT3AMT has been studied mostly in AML,1,2 and only a few studies compared the clinical impact of various types of DNMT3AMT, their position within the clonal hierarchy (primary vs secondary), and their configuration (canonical vs non-canonical DNMT3AMT).3
Heterozygous DNMT3AMT are most commonly found in noncore binding factor (NCBF) AML, but also occur in MDS and related neoplasms.1,4–6 Subclonal DNMT3AMT were found in otherwise asymptomatic older individuals and linked to increased risk of subsequent leukemia.7,8 While most common DNMT3AMT affect the canonical site (R882), nonsense variants have also been reported, but 2p deletions or microdeletions affecting the DNMT3A locus are rare.9 The canonical R882 profoundly reduces methyltransferase activity, suggesting a dominant-negative effect,10 whereas isolated nonsense mutations produce haploinsufficiency. The lack of somatic uniparental disomy, hemizygous deletions, or biallelic mutations suggests that such lesions would be non-competitive/lethal to the clone.
We investigated the role of mutation topography, clonal hierarchical position and clinical features in MDS, overlap neoplasms (MPN and MDS/MPNs), and primary and secondary AML to elucidate the clinical characteristics of DNMT3AMT cases with respect to molecular biology and clinical phenotypes.
Mutational status was analyzed in 422 MDS, 240 MDS/MPN, 163 sAML and 234 pAML cases (n = 1059) followed for 1–311 mo. (median 53 mo.; Supplementary Table S1, Supplementary Figure S1A). Among them, 155 samples were subjected to whole-exome sequencing and 904 tested for mutations in 62 genes using targeted deep NGS (Supplementary Tables S2 and S3). Somatic DNMT3AMT were identified in 212 (21%) patients (Table 1, Figure 1a). Alterations predicted to have functional consequences based on ≥ 5/9 databases are shown in Supplementary Table S4. Of the 212 mutations, 150 were missense (78 canonical R882, 72 non-canonical) and 62 truncations/frame shifts (Figure 1a) that were all heterozygous; 17/1059 patients had germline variants of unknown significance (Supplementary Table S5).
Table 1.
Patient characteristics among different variants in DNMT3A mutation
| Total Cohort | DNMT3A WT | DNMT3A MT | R882 | Truncating | NCMS | |
|---|---|---|---|---|---|---|
| n | 1059 | 847 | 212 | 78 | 62 | 72 |
| Age (median) | 65 | 60 | 66* | 67 | 68 | 67 |
| Gender % (M) | 58 | 58 | 56 | 56 | 55 | 60 |
| Diagnosis | ||||||
| MDS | 422 | 347 (82%) | 75 (18%) | 17 (4%) | 25 (6%) | 33 (8%) |
| pAML | 234 | 152 (65%) | 82 (35%)** | 47 (20%)*** | 18 (8%) | 17 (7%) |
| sAML | 163 | 133 (82%) | 30 (18%) | 8 (5%) | 9 (6%) | 13 (8%) |
| MDS/MPN | 240 | 215 (90%) | 25 (10%) | 6 (3%) | 10 (4%) | 9 (4%) |
| Normal karyotype | 434 (51%) | 85 (40%) | 32 (41%) | 20 (32%) | 33 (45%) | |
| Complex karyotype | 116 (14%)**** | 14 (6%) | 5 (6%) | 4 (6%) | 5 (7%) | |
| CBC (mean) | ||||||
| WBC × 109/l | 4 | 5 | 4 | 5.6 | 6.4 | |
| ANC × 109/l | 2 | 2 | 2 | 2.3 | 2.7 | |
| Hb (g/dl) | 10 | 9 | 10 | 9.6 | 9.6 | |
| Platelet count (k/μl) | 81 | 44 | 36 | 63 | 33 | |
| PB Blast % | 3 | 12 | 23***** | 10 | 6 | |
| BM blast % | 10 | 18 | 30***** | 18 | 10 |
Abbreviations: ANC, absolute neutrophil count; BM, bone marrow; CBC, complete blood count; Hb, hemoglobin; MDS/MPN, Myelodysplastic syndrome/Myeloproliferative neoplasm; NCMS, non-canonical missense mutation in DNMT3A gene; pAML, primary AML; PB, peripheral blood; sAML, secondary AML; WBC, white blood cells.
P < 0.001 between DNMT3AMT vs wild type;
P < 0.001 comparing prevalence of DNMT3AMT among MDS, pAML, sAML and MDS/MPN;
P < 0.001 between R882 vs Truncating vs other non-canonical missense DNMT3AMT;
P < 0.05 between DNMT3AMT vs wild type;
P = 0.05 between R882 DNMT3AMT vs DNMT3AWT.
Figure 1.
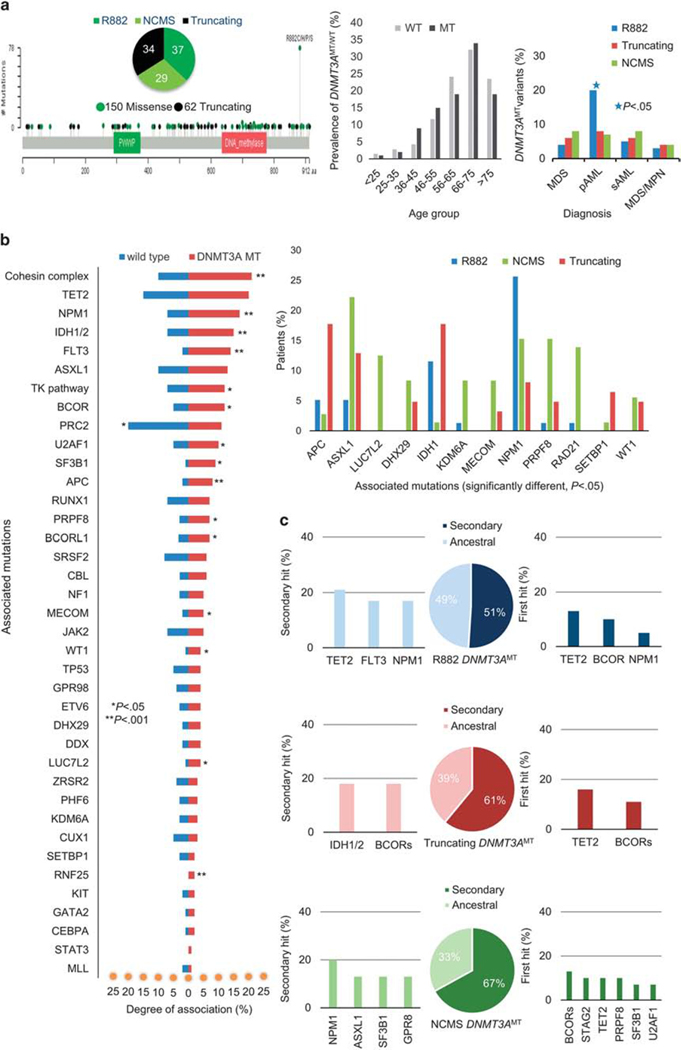
Characteristics of DNMT3AMT in our cohort of patients with myeloid neoplasms. (a) Illustration of position of DNMT3AMT in chromosome 2 in our cohort of patients with myeloid neoplasia (left). Prevalence of DNMT3AMT in different age groups in comparison to DNMT3AWT (center). Maximum prevalence of DNMT3AMT between age group 66–75 years. Distribution of different DNTMT3AMT variants among different myeloid malignancies in the whole cohort (right). Canonical R882 variant is strongly associated with pAML than truncating or NCMS DNMT3AMT (*P < 0.05). (b) ‘Tornado chart’ (left) demonstrating associated mutations in DNMT3AMT and in wild-type patients with different myeloid neoplasia. Bar graph (right) shows significantly different other associated myeloid mutations in R882, NCMS and truncating DNMT3AMT variants. There is a higher association of NPM1 in R882 variant, APC, IDH1 and SETBP1 in truncating variant and ASXL1, PRPF8 and RAD21 in NCMS DNMT3AMT. (c) Illustrates clonal architecture in R882, truncating and NCMS variants of DNMT3AMTmyeloid neoplasms.
DNMT3AMT were most common in pAML (35%), followed by MDS or sAML (each 18%) and MDS/MPN (10%; P < 0.000001; Table 1, Supplementary Figure S1B). DNMT3AMT were associated with older age compared to DNMT3AWT (66 vs 60 y; P = 0.0001; Table 1, Figure 1a). R882 DNMT3AMT coincided with pAML more than truncating or NCMS (non-canonical missense) variants (60 vs 29 vs 24%; P = 0.004; Figure 1a). R882 was, however, under-represented in MDS compared to non-R882 (22 vs 43%, P = 0.03; Table 1, Supplementary Figures S2A–D). Complex karyotypes were less frequent in DNMT3AMT vs DNMT3AWT cases (6 vs 14%; P = .009; Table 1).
Compared to DNMT3AWT, DNMT3AMT coincided with certain additional hits (Figure 1b), including those in cohesin genes, NPM1, IDH1/2, and FLT3. In contrast, PRC2 complex mutations (SUZ12/EED/EZH2) associated more with DNMT3AWT than with DNMT3AMT. Among DNMT3AMT types, NPM1 mutations were found more with R882 than with truncating and NCMS DNMT3AMT (26 vs 8 vs 15%, P = 0.001). APC and IDH1 were mutated more in truncating than in NCMS or R882 DNMT3AMT (18 vs 3 vs 5%, P = .01 and 18 vs 1 vs 12%, P = 0.006). ASXL1 (22 vs 13 vs 5%, P = .02), PRPF8 (15 vs 5 vs 1%, P = 0.007), RAD21 (14 vs 0 vs 1%, P = 0.0006) and LUC7L2 (13% vs 0 vs 0, P = 0.0001) were frequently co-mutated with NCMS (Figure 1b).
Analyzing DNMT3AMT in AML, pAML patients were more likely to harbor NPM1, FLT3 and IDH1 mutations than sAML (28 vs 0%, P = 0.002; 23 vs 3%, P = 0.04; 17 vs 3%, P = 0.1, respectively). Mutations in ASXL1 and SF3B1 more frequently coincided with MDS and sAML vs. pAML (15 vs 5%, P = 0.05; 12 vs 1%, P = 0.008; Supplementary Figure S3).
In a clonal hierarchy analysis to determine whether DNTM3AMT were ancestral or secondary genetic events (Supplementary Table S6, Figure 1c, Supplementary Figure S4), the median VAF (variant allele frequency) for DNMT3AMT cases was 35% (3–93%). Median VAFs among DNMT3AMT types may differ at 41, 35 and 27% in R882, truncating, and NCMS DNMT3AMT, respectively (Supplementary Figure S5). DNMT3AMT was ancestral in 42% of cases. In these, the most common second hits were in TET2, NPM1, FLT3, and IDH1/2 (Supplementary Figure S4A). Approximately ½ (49%) of R882 variants were ancestral vs 1/3 of non-R882 variants (39% truncating, 33% NCMS, P = 0.04). The most common secondary hits in dominant R882 DNTM3AMT cases were TET2, FLT3 and NPM1, while in dominant truncating DNMT3AMT cases, BCOR and IDH1/2 variants were the most common. In dominant NCMS DNMT3AMT cases, mutations in NPM1, ASXL1 and SF3B1 were the most common second hits (Figure 1c). TET2 was a common ancestral hit associated with DNMT3AMT (Supplementary Figure S4). Among the DNMT3AMTcases (R882 and truncating mutations), TET2 was the most frequent ancestral event (Figure 1c).
DNMT3AMT associated with worse overall survival (OS) than DNMT3AWT (OS plots by univariate analyses) (Supplementary Figure S6A), failing to reach significance only in MDS/MPN and low-risk MDS, wherein DNMT3AMT vs DNMT3AWT median OS was 37 vs 102 mo. (P = 0.12) and 69 vs 101 mo. (P = 0.54), respectively (Supplementary Figures S6B, C); in high-risk MDS and pAML +sAML, median OS were 26 vs 75 mo. (P < 0.0001; Supplementary Figure S6D) and 16 vs 76 mo. (P < 0.0001) (Supplementary Figure S6E), respectively. There was no significant difference in OS in R882 vs NCMS or truncating DNMT3AMT (26 vs 23 vs 27 months; P = 0.72; Supplementary Figure S6F). There was also no significant OS difference for patients with ancestral vs. secondary DNMT3AMT (28 vs 17 mo., P = 0.2; Supplementary Figure S6G). Of note is that DNMT3AMT with normal cytogenetics had better OS than those with abnormal/complex cytogenetics (32 vs 16 mo., P = 0.001; Supplementary Figure S6H). We also did a multivariate additive effects Cox model analysis which yielded hazard ratios (HR) relative to DNMT3AWT low-risk MDS (Supplementary Table S7). These results show that assuming additive effects, the impact on survival of disease classification dwarfs the impact of different types of DNMT3AMT, but given additional interaction model parameters and thus flexibility in data fitting, DNMT3AMT type main effects differ and are strongest for R882 (P = 0.03).
DNMT3A mutation is considered an early lesion with leukemogenic potential.11 By itself, DNMT3AMT may be insufficient to cause neoplasia, as seen in asymptomatic older carriers7,8 and in heterozygous murine models.12 Our results demonstrate that clonal configuration and mutation type contribute to the heterogeneity of clinical outcomes and explain the difficulty in precisely assigning prognoses to a DNMT3AMT. As in previous reports,1,2,10 we identified these lesions in up to 35% of AML patients with canonical DNMT3AR882 being most common. The acuity of this disease suggests that R882 has the strongest leukemogenic potential. These findings are consistent with the dominant-negative role of DNMT3AR882 variant causing a degree of dysfunction beyond haploinsufficiency.10 Indeed, R882 caused greater reductions in methyltransferase activity than other heterozygous hits.13 In MDS/related disorders, DNMT3A truncations and NCMS variants predominate, implying that they associate with a more protracted disease. Likely, the non-canonical DNMT3A variants are less proleukemic, and require additional myeloid mutations.
The notion that DNMT3AR882 is more virulent is supported by the fewer secondary events. Also, as a secondary hit, DNMT3AR882 rapidly ‘sweeps’ the entire clonal population, thus requiring few other associated lesions. In contrast, non-R882 and truncating mutations may not suffice without other hits. The difference in associated mutations among the three variants reinforces the notion that R882 DNMT3AMT has a dominant-negative impact. It also suggests that each variant has distinct biological features.
The association of DNMT3AMT with aggressive diseases is difficult to reconcile with the presence of subclonal founder lesions in asymptomatic, older adults.7,8 This raises the question as to whether these asymptomatic carriers preferentially develop MDS, rather than pAML and/or are more likely to be affected by the less potent and non-canonical or truncation variants. Indeed, most of the DNMT3A clones reported in healthy, older individuals were not R882 (only 12–17% of healthy elderly DNMT3AMT were R882).7,8 Our results show that the spectrum of variants present in MDS is similar to that in asymptomatic carriers, consistent with the older age of onset of MDS. Consequently, additional hits may be needed to induce frank clonal neoplasia.
Using targeted gene panels, we reconstructed the clonal architecture of individual cases within disease subtypes. DNMT3AMT were more often ancestral in pAML, whereas they can also be secondary subclonal in MDS and post-MDS (secondary) AML. In general, ancestral DNMT3AMT or TET2MT in MDS imply that such cases may have originated from clonal hematopoiesis of indeterminate potential (CHIP) of older adults. In contrast, other ancestral hits not observed in CHIP or subclonal DNMT3AMT may point toward other etiologies. Nevertheless, truncations and NCMS DNMT3A lesions in MDS, which could be derived from CHIP, more likely represent secondary lesions. R882 is mostly a founder lesion, irrespective of histology, and is often followed by phenotype-determining mutations in NPM1, FLT3, or IDH1, which is consistent with murine models.14
The rank of the DNMT3AMT in clonal hierarchy may have therapeutic implications. Ancestral DNMT3AMT may be more likely to persist in relapses after treatments with IDH1/2 or FLT3 inhibitors. In contrast, secondary DNMT3A hits following IDH1/2 or other targetable defects may be easier to eradicate. DNMT3AMT confers a poor prognosis across different myeloid neoplasms. In terms of their position in the clonal hierarchy, no difference in outcomes between ancestral and secondary DNMT3A hits were observed which is consistent with DNMT3AMT being clone sweeping events.15
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by the EPE1509JM - Edward P Evans Foundation, R01HL123904 – NHLBI, R01HL118281 – NHLBI, R01HL128425 – NHLBI.
Footnotes
Supplementary Information accompanies this paper on the Leukemia website (http://www.nature.com/leu)
CONFLICT OF INTEREST
The authors declare no conflict of interest.
REFERENCES
- 1.Ley TJ, Ding L, Walter MJ, McLellan MD, Lamprecht T, Larson DE et al. DNMT3A mutations in acute myeloid leukemia. N Engl J Med 2010; 363: 2424–2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ribeiro AFT, Pratcorona M, Erpelinck-Verschueren C, Rockova V, Sanders M, Abbas S et al. Mutant DNMT3A: a marker of poor prognosis in acute myeloid leukemia. Blood 2012; 119: 5824–5831. [DOI] [PubMed] [Google Scholar]
- 3.Roller A, Grossmann V, Bacher U, Poetzinger F, Weissmann S, Nadarajah N et al. Landmark analysis of DNMT3A mutations in hematological malignancies. Leukemia 2013; 27: 1573. [DOI] [PubMed] [Google Scholar]
- 4.Shih AH, Abdel-Wahab O, Patel JP, Levine RL. The role of mutations in epigenetic regulators in myeloid malignancies. Nat Rev Cancer 2012; 12: 599–612. [DOI] [PubMed] [Google Scholar]
- 5.Walter MJ, Ding L, Shen D, Shao J, Grillot M, McLellan M et al. Recurrent DNMT3A mutations in patients with myelodysplastic syndromes. Leukemia 2011; 25: 1153–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abdel-Wahab O, Pardanani A, Rampal R, Lasho T, Levine R, Tefferi A. DNMT3A mutational analysis in primary myelofibrosis, chronic myelomonocytic leukemia and advanced phases of myeloproliferative neoplasms. Leukemia 2011; 25: 1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Genovese G, Kähler AK, Handsaker RE, Lindberg J, Rose SA, Bakhoum SF et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N Engl J Med 2014; 371: 2477–2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jaiswal S, Fontanillas P, Flannick J, Manning A, Grauman PV, Mar BG et al. Age-related clonal hematopoiesis associated with adverse outcomes. N Engl J Med 2014; 371: 2488–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang L, Rau R, Goodell MA. DNMT3A in haematological malignancies. Nat Rev Cancer 2015; 15: 152–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Russler-Germain DA, Spencer DH, Young MA, Lamprecht TL, Miller CA, Fulton R et al. The R882H DNMT3A mutation associated with AML dominantly inhibits wild-type DNMT3A by blocking its ability to form active tetramers. Cancer Cell 2014; 25: 442–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xie M, Lu C, Wang J, McLellan MD, Johnson KJ, Wendl MC et al. Age-related mutations associated with clonal hematopoietic expansion and malignancies. Nat Med 2014; 20: 1472–1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Challen GA, Sun D, Jeong M, Luo M, Jelinek J, Berg JS et al. DNMT3A is essential for hematopoietic stem cell differentiation. Nat Genet 2012; 44: 23–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Holz-Schietinger C, Matje DM, Reich NO. Mutations in DNA methyltransferase (DNMT3A) observed in acute myeloid leukemia patients disrupt processive methylation. J Biol Chem 2012; 287: 30941–30951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim SJ, Zhao H, Hardikar S, Singh AK, Goodell MA, Chen T. A DNMT3A mutation common in AML exhibits dominant-negative effects in murine ES cells. Blood 2013; 122: 4086–4089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Makishima H, Yoshizato T, Yoshida K, Sekeres MA, Radivoyevitch T, Suzuki H et al. Dynamics of clonal evolution in myelodysplastic syndromes. Nat Genet 2016; 49: 204–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.