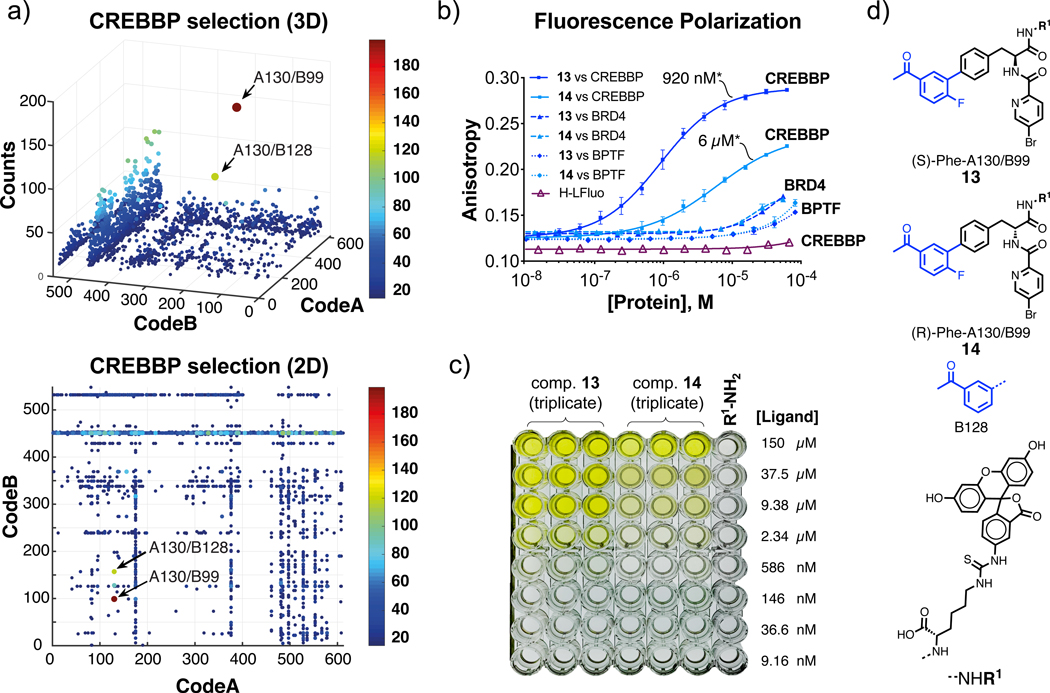
Figure 3 |. Selection against CREBBP bromodomain, hit validation and selectivity determination.
a, Fingerprint of a CREBBP bromodomain selection plotted in 3D and 2D representation, respectively. The x and y axes represent the two building blocks while the color heat map and z-axis correspond to the DNA sequence counts. A cut-off value equal to 15 was applied to fingerprints. Combinations A130/B99 and A130/B128 were preferentially enriched (EFA130/B99=149±16 and EF A130/B128=76±3. For the calculation of EFs see Supplementary Tables 2,3 and Equation 1 (S29). Selections were performed in triplicate (see Supplementary Figure 21). b, Fluorescence polarization of FITC-labelled compounds 13 and 14 with CREBBP bromodomain and control bromodomains BRD4(1) and BPTF. Error bars indicate standard deviation of three measurements.
The asterisk (*) indicates the dissociation constant value (Kd). c, Snapshot of maxisorp plate carrying small-molecule ELISA of compounds 13 and 14 after addition of TMB and 1M H2SO4. The assay was performed in triplicate against immobilized CREBBP bromodomain. d, Chemical structures of selected compounds and building blocks.