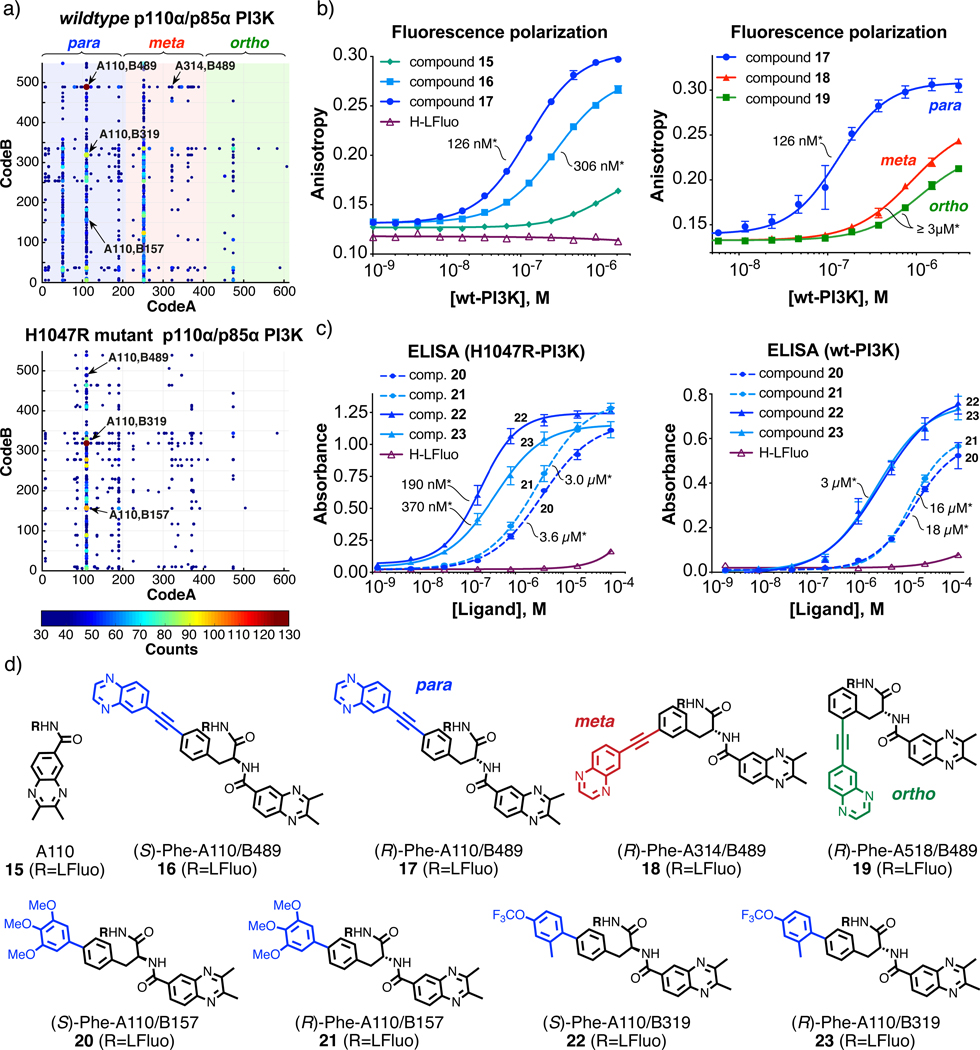
Figure 4 |. Selections against PI3KCA variants.
a, Two-dimensional fingerprints of NF-DEL selections against the PI3Ks wildtype p110α/p85α PI3K (upper panel) and H1047R-p110α mutant of p110α/p85α PI3K (lower panel). The x and y axes represent the two building blocks while the color heat map shows the HTDS sequence counts. Building blocks A, corresponding to para- (1–204), meta- (205–408) and ortho- (409–612) regio-isomers, are highlighted in blue, red and green, respectively. Cut-off values equal to 25 (for wt-PI3K variant selection) and 30 (for H1047R mutant of PI3K variant selection) were applied to the fingerprints. The most enriched building block combinations are indicated with arrows. For the calculation of EFs see Supplementary Tables 2,3 and Equation 1 (S29). b, The binding of the most enriched combination (A110/B489, EF=65±7) against the wildtype protein corresponding to stereoisomers 15 and 16 was validated by fluorescence polarization and compared with a derivative of building block A110 alone (15). The meta- and ortho-isomers of 17 corresponding to combinations A314/B489 (18) and A518/B489 (19) were also measured for comparison. Error bars indicate standard deviation of three measurements. The asterisk (*) indicates the dissociation constant value (Kd). c, Enriched compounds corresponding to combinations A110/B157 (20 and 21) and A110/B319 (22 and 23) were validated by small-molecule ELISA against both H1047R mutant (left panel) and wildtype PI3K (right panel). Error bars indicate standard deviation of three measurements. The asterisk (*) indicates the dissociation constant value (Kd). d, Chemical structures of selected compounds.