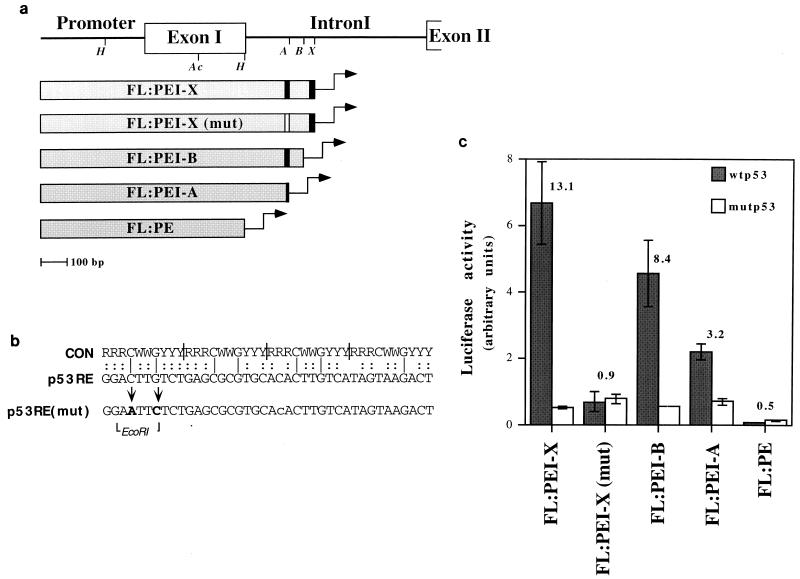
FIG. 4.
Deletion analysis mapping of the region responsible for p53-mediated c-fos transactivation. (a) Schematic map of the reporter plasmids used for transactivation analysis. Each plasmid contains the indicated region of the c-fos gene placed upstream of the luciferase gene in a promoterless luciferase construct (pGL3basic). Italics identify the positions of restriction enzymes used to create the various deletion mutants: H, HincII; Ac, AccI; A, ApaLI; B, BsmI; X, XhoI. Construct FL:PE was created through partial digestion of FL:PEI with HincII. The black vertical bars indicate the two clusters of p53 consensus half sites (Fig. 3a). The white bar represents the mutated p53-RE. (b) Alignment of the consensus (CON) p53-binding site (12) and the 40-bp p53-RE of the mouse c-fos gene. Vertical bars within the consensus sequence indicate the four half-site decamers. The positions of the site-directed mutations in the p53-RE, as well as that of the unique EcoRI site created during mutagenesis, are shown at the bottom. (c) Luciferase activity of the reporter plasmids depicted in panel a. For details, see the legend to Fig. 3c.