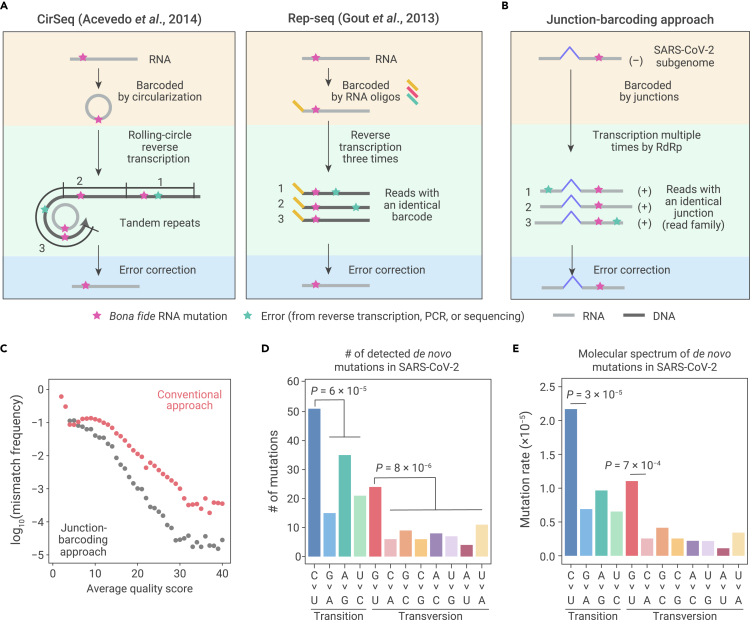
Figure 1.
The molecular spectrum of de novo SARS-CoV-2 mutations
(A) Schematic of two experimental approaches previously developed to detect RNA mutations. Bona fide RNA mutations (magenta stars) should be repeatedly detected, while errors generated during reverse transcription, PCR amplification, or high-throughput sequencing (green stars) should only be occasionally detected.
(B) Schematic of our junction-barcoding approach to detect RNA mutations for SARS-CoV-2. The genomic coordinates of a pair of upstream and downstream sites of sporadic junctions can serve as the molecular barcode to group sequencing reads derived from the same negative-sense subgenome into read families. Bona fide RNA mutations should be unanimously detected in a read family.
(C) Comparison of overall mismatch frequency between our junction-barcoding approach and the conventional computational approach.
(D) The numbers of de novo RNA mutations of 12 base-substitution types, with respect to the positive-sense SARS-CoV-2 genome. Two-tailed p values were calculated from binomial tests assuming an equal frequency for each type of base substitutions.
(E) The molecular spectrum of de novo SARS-CoV-2 mutations. Two-tailed p values were calculated from Fisher's exact tests.