Figure 2.
The molecular spectrum of SARS-CoV-2 polymorphisms among patients
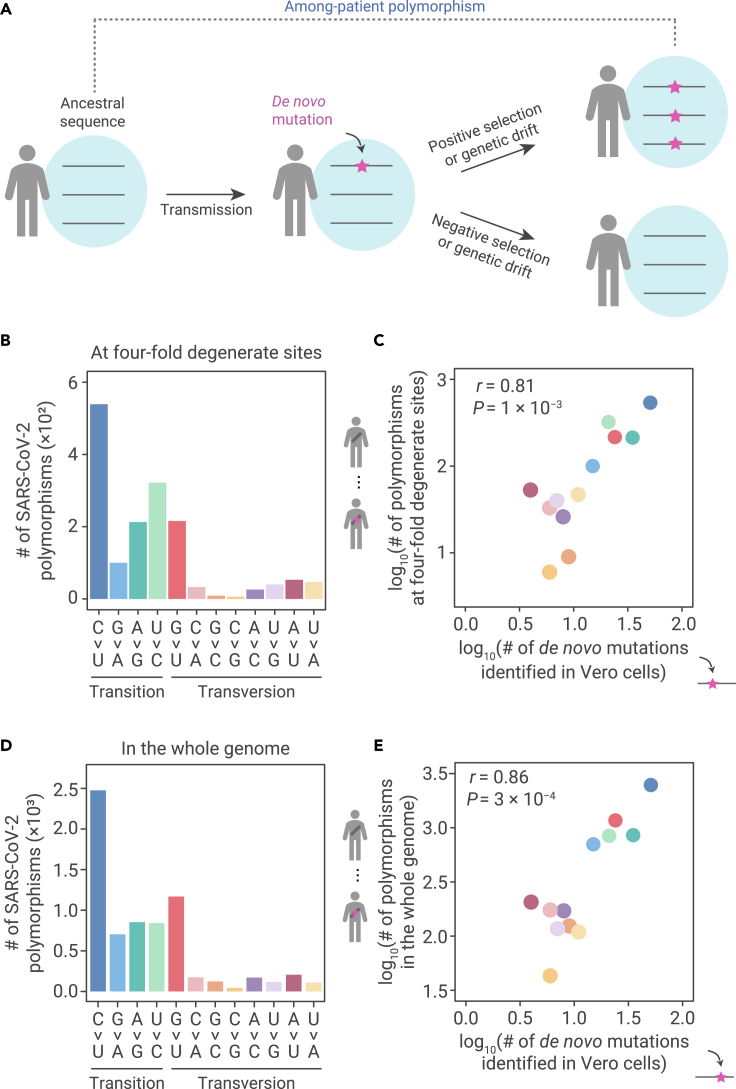
(A) The emergence of among-patient polymorphisms through the accumulation of de novo mutations. The frequency of a de novo mutation (the magenta star with an arrow pointing to it) may be increased by positive selection, decreased by negative selection, or changed through genetic drift due to chance events. If a mutation becomes predominant within a patient, it can be detected as an among-patient polymorphism.
(B) The molecular spectrum of among-patient polymorphisms at 4-fold degenerate sites in SARS-CoV-2.
(C) A scatterplot shows the molecular spectrum of de novo mutations versus among-patient polymorphisms at 4-fold degenerate sites in SARS-CoV-2. Each dot represents a base-substitution type, colored according to (B). Pearson’s correlation coefficient (r) and the corresponding p value are shown.
(D) The molecular spectrum of among-patient polymorphisms in the whole genome of SARS-CoV-2.
(E) Similar to (C), for all polymorphisms.