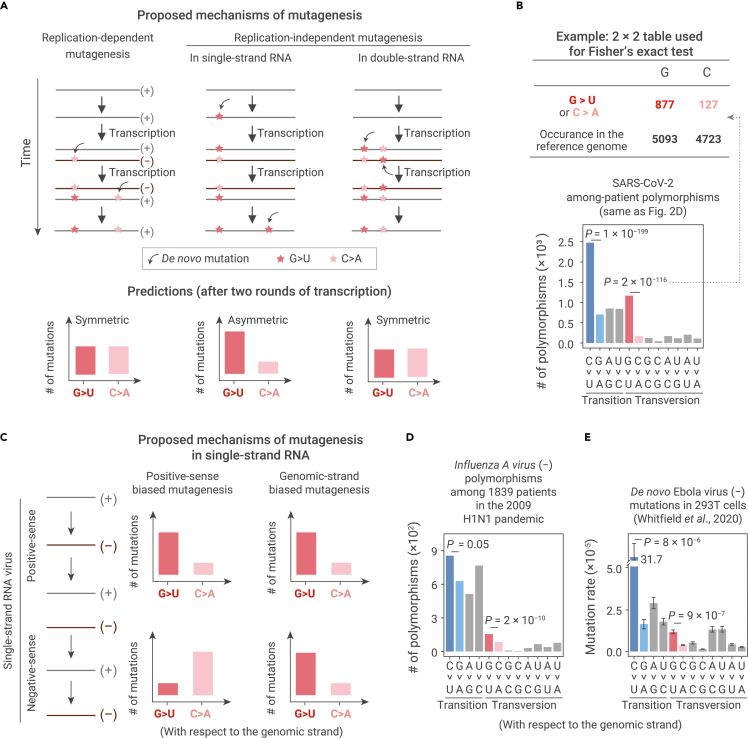
Figure 3.
Predictions and observations for various mutagenic mechanisms on the symmetry of mutations
(A) Predictions on the symmetry between a pair of complement base-substitution types for three potential mutagenic mechanisms. If de novo mutations are introduced during transcription by RdRp (left panel), or by a replication-independent mechanism in double-strand RNAs (right panel), mutations should be symmetric when a replication cycle is completed: a base-substitution type and its complement base-substitution type should arise at the same rate in the viral genome. On the contrary, if de novo mutations are introduced by a replication-independent mechanism specific to single-strand RNAs, mutations could be asymmetric (middle panel).
(B) The statistical assessment on the symmetry of mutations using Fisher's exact tests.
(C) Predictions for two potential mutagenic mechanisms in single-strand RNAs, positive-sense biased versus genomic-strand biased mutagenesis.
(D) The molecular spectrum of among-patient polymorphisms in a negative-sense, single-strand RNA virus, Influenza A virus (subtype H1N1). Two-tailed p values were calculated from Fisher's exact tests.
(E) The molecular spectrum of de novo mutations in a negative-sense, single-strand RNA virus, Ebola virus. De novo mutations were identified from isolated virions, at which time replication cycles have completed. Error bars represent standard errors (N = 21) of the average mutation rates of each base-substitution type. Two-tailed p values were calculated using the t tests.