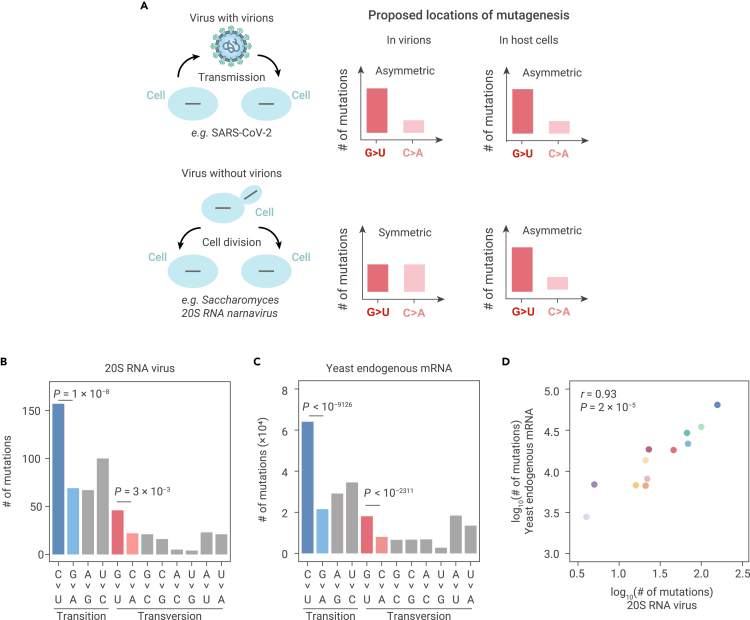
Figure 4.
Predictions and observations for mutagenic processes in virions versus in host cells
(A) Predictions on the symmetry of mutations for mutagenic processes in virions versus in host cells.
(B) The molecular spectrum of de novo mutations that we detected in 20S RNA narnavirus from previously published ARC-seq data. Two-tailed p values were calculated from Fisher's exact tests.
(C) The molecular spectrum of yeast mRNA mutations that we detected from previously published ARC-seq data. Two-tailed p values were calculated from Fisher's exact tests.
(D) A scatterplot shows the molecular spectrum of de novo mutations in 20S RNA narnavirus versus in yeast endogenous mRNAs. Each dot represents a base-substitution type, colored according to Figure 1E. Pearson's correlation coefficient (r) and the corresponding p value are shown.