Abstract
Background
Multisystem inflammatory syndrome in children (MIS-C) consensus criteria were designed for maximal sensitivity and therefore capture patients with acute COVID-19 pneumonia.
Methods
We performed unsupervised clustering on data from 1,526 patients (684 labeled MIS-C by clinicians) <21 years old hospitalized with COVID-19-related illness admitted between 15 March 2020 and 31 December 2020. We compared prevalence of assigned MIS-C labels and clinical features among clusters, followed by recursive feature elimination to identify characteristics of potentially misclassified MIS-C-labeled patients.
Findings
Of 94 clinical features tested, 46 were retained for clustering. Cluster 1 patients (N = 498; 92% labeled MIS-C) were mostly previously healthy (71%), with mean age 7·2 ± 0·4 years, predominant cardiovascular (77%) and/or mucocutaneous (82%) involvement, high inflammatory biomarkers, and mostly SARS-CoV-2 PCR negative (60%). Cluster 2 patients (N = 445; 27% labeled MIS-C) frequently had pre-existing conditions (79%, with 39% respiratory), were similarly 7·4 ± 2·1 years old, and commonly had chest radiograph infiltrates (79%) and positive PCR testing (90%). Cluster 3 patients (N = 583; 19% labeled MIS-C) were younger (2·8 ± 2·0 y), PCR positive (86%), with less inflammation. Radiographic findings of pulmonary infiltrates and positive SARS-CoV-2 PCR accurately distinguished cluster 2 MIS-C labeled patients from cluster 1 patients.
Interpretation
Using a data driven, unsupervised approach, we identified features that cluster patients into a group with high likelihood of having MIS-C. Other features identified a cluster of patients more likely to have acute severe COVID-19 pulmonary disease, and patients in this cluster labeled by clinicians as MIS-C may be misclassified. These data driven phenotypes may help refine the diagnosis of MIS-C.
Funding
This work was funded by the US Centers for Disease Control and Prevention (75D30120C07725) and National Institutes of Health (K12HD047349 and R21HD095228).
Keywords: COVID-19, Multisystem inflammatory syndrome, Pediatrics, Critical care medicine, Clustering
Research in context.
Evidence before this study
Consensus criteria designed for disease surveillance for multisystem inflammatory syndrome in children (MIS-C) emphasized sensitivity over specificity and encompass some patients with acute COVID-19 multisystem involvement. We performed a search in PubMed on 9 April 2021 using the terms “multisystem inflammatory syndrome in children” or “pediatric multisystem inflammatory syndrome” or “pediatric inflammatory multisystem syndrome.” Of 58 case series or cohort studies with ≥20 patients meeting criteria for MIS-C (only six included ≥100 patients), two studies applied an unbiased data-driven approach to clinical features to refine the phenotype: one focused solely on biomarkers; the other included only patients diagnosed with MIS-C and did not compare them to non-MIS-C-diagnosed COVID-19 patients.
Added value of this study
We used an unbiased, unsupervised clustering analysis to group 1526 patients hospitalized with COVID-19-related complications, including 684 labeled MIS-C, into subphenotypes with distinct clinical and biological profiles and identified three distinct subphenotypes: 1) hyperinflamed patients who were SARS-CoV-2 antibody positive and RT-PCR negative, had cardiovascular involvement and/or Kawasaki-like features, and were often critically ill requiring vasoactive infusions; 2). a respiratory cluster, 90% of whom were SARS-CoV-2 RT-PCR positive, 79% of whom had pulmonary infiltrates, and 36% of whom required mechanical ventilation with the majority of patients having underlying health conditions; and 3) patients who had a relatively mild course of illness and rarely required critical care. While 92% of patients in the first cluster were clinically labeled as having MIS-C, nearly 30% of the patients in the respiratory cluster were labeled as having MIS-C and were treated with both antiviral and immunomodulatory therapies.
Implications of all the available evidence
These findings suggest there is a prototypical MIS-C phenotype that includes severely ill, hyperinflamed patients with cardiovascular involvement and/or Kawasaki disease-like features. Certain laboratory, clinical, and demographic features are helpful in classifying patients as MIS-C vs. acute COVID-19. The subgroup of critically ill, SARS-CoV-2 RT-PCR positive patients with pulmonary infiltrates and severe cardiovascular involvement may be a distinct phenotype from prototypical MIS-C patients meriting a different therapeutic approach.
Alt-text: Unlabelled box
1. Introduction
In April 2020, a severe illness in children and adolescents temporally associated with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection, with features of Kawasaki disease [1], [2], [3] (KD) or cardiovascular shock, was reported [4]. Criteria for this new illness [5], [6], [7], [8], [9]called multisystem inflammatory syndrome in children (MIS-C) by the US Centers for Disease Control and Prevention (CDC) and pediatric inflammatory multisystem syndrome temporally associated with SARS-CoV-2 (PIMS-TS) in Europe and the United Kingdom [10,11]were published as online health alerts [12,13]. CDC criteria for MIS-C include hospitalization, fever, elevated laboratory markers of inflammation, involvement of at least two organ systems, and evidence of exposure to SARS-CoV-2 [12]. These criteria, based on limited data from early case series, were rapidly published for public health tracking [1,4]. With incomplete understanding of disease pathophysiology and its temporal course, MIS-C criteria were necessarily broad [14] and likely overlap with those of severe coronavirus disease 2019 (COVID-19), which is increasingly reported in young individuals [15], [16], [17], [18]. Acute respiratory distress syndrome (ARDS), common in critically ill patients with COVID-19,[17] is often associated with hyperinflammation [19] and multiorgan dysfunction with features of septic shock, similar to MIS-C. Some authors have recently suggested framing these syndromes along a continuum of disease that includes Acute COVID-19 Cardiovascular Syndrome in adults, in whom the cardiovascular phenotype can present contemporaneously with pneumonia.[20]
We sought to discover distinct subphenotypes, including MIS-C, within a cohort of patients with COVID-19-related illness using unsupervised (i.e., not based on clinician-assigned labels of MIS-C), data-driven methods. Our hypothesis was that a machine learning approach would differentiate patients with MIS-C from severe acute COVID-19 and milder disease. We also hypothesized that clinician-assigned labels of MIS-C would span multiple subphenotypes, demonstrating the heterogeneity of this syndromic definition. Finally, we aimed to identify specific features that differentiate MIS-C from severe COVID-19 respiratory illness.
2. Methods
2.1. Design and cohort
Data on children and adolescents hospitalized with COVID-19-related symptoms from the Overcoming COVID-19 National Public Health Surveillance Registry [5] were analyzed using a machine-learning, unsupervised clustering approach. Data were abstracted by trained research staff on patients admitted between 15 March 2020 and 31 December 2020. Inclusion criteria were: 1) hospitalization with symptoms related to COVID-19; 2) age <21 years; 3) positive SARS-CoV-2 test during illness (reverse transcriptase polymerase chain reaction (RT-PCR) or antibody) or, if MIS-C, exposure to a person with COVID-19. The study was conducted in accordance with the Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) guidelines and a STROBE checklist is included with the manuscript (see Supplementary) [21]. The registry was determined by the institutional review board at Boston Children's Hospital to meet the requirement of public health surveillance with waiver of informed consent and conducted consistent with applicable federal law and CDC policy.
2.2. Variable definitions and data processing
Positive anti-SARS-CoV-2 antibody results were defined by individual laboratories’ criteria. Specific medication and drug classes included in the analysis are outlined in the appendix (pp. 6–7). Patients were labeled as having MIS-C by clinician investigators at each site, with secondary confirmation that each patient met CDC criteria for MIS-C by registry principal investigators (MMP, AGR) [5]. All variables with repeated evaluation during a patient's hospitalization were summarized using the worst (minimum or maximum) value (appendix, pp. 89). Additional variable definitions are provided in the appendix (pp. 6,7).
2.3. Unsupervised clustering
Our clustering approach identifies patterns in data that minimize separation within clusters and maximize separation between clusters, without reliance on a predetermined outcome variable to train the algorithm (appendix, p. 11). We did not use clinician-assigned MIS-C labels to train the clustering algorithm. This method avoids investigator biases that may influence traditional classification methods, such as logistic regression, which use positively and negatively labeled patients to train models.
Initial inputs to the clustering algorithm were 94 variables (appendix, pp. 8,9). We did not include anti-SARS-CoV-2 serology as an input variable because it was not measured in the majority (61%) of patients who had otherwise complete data and because anti-SARS-CoV-2 antibody testing was not broadly available early in the registry period. Troponin was not included due to assay variability across clinical site laboratories [22]. Whether various biomarkers were measured for each patient was at the discretion of the treating clinicians. Therefore, when laboratory values were unmeasured (“missing”), this missingness was not at random, and thus indicator variables for whether each laboratory value was measured were included in the model.
We used all candidate variables as initial inputs to a partitioning around medoids (PAM) clustering algorithm. An initial cluster assignment was recorded for each patient. Backwards selection using a random forest classifier with cluster as the target class was used to determine the optimal number of features needed for clustering. We then reapplied the clustering algorithm using only the selected features to assign final cluster membership to each patient. Details of data preparation, variable selection, and clustering are in the appendix (p. 4).
2.4. Cluster evaluation
Bootstrapping with 1000 clustering replications was used to determine the precision around the number and percent of patients in each cluster with an MIS-C label and with other clinical features. Given the exploratory nature of this analysis, multiple comparisons, and challenges in inference testing with variables used for clustering [23], we report the spread around point estimates among the 1000 replications rather than p-values for statistical tests. Cluster stability was assessed by counting the proportion of times a pair of patients clustered together divided by the number of times the pair of patients was selected in a bootstrap resample. Ideal clustering should result in each pair of patients always or never clustering together (appendix, p. 4).
To evaluate the relative importance of cluster assignment and MIS-C labels, we fit univariable and multivariable logistic regression models using cluster membership and MIS-C diagnosis as independent variables and a composite critical illness outcome as the dependent variable. Critical illness was defined as at least one of the following: (1) death; (2) extracorporeal membrane oxygenation (ECMO); (3) vasoactive infusions; (4) invasive or noninvasive mechanical ventilation; or (5) new initiation of dialysis. We used C-statistics to determine model discrimination and applied three methods to evaluate which variable most contributed to models’ overall discrimination. First, we compared p-values obtained from likelihood ratio tests comparing the multivariable model to each univariable model with only cluster or MIS-C as independent variables. Second, we compared C-statistics between univariable models. Finally, we used dominance analysis [24], with Nagelkerke's coefficient of determination as the measure of fit [25], to determine variable importance. Furthermore, we verified these results by using recursive feature elimination to determine feature importance using a random forest classifier (appendix, p. 4) with critical illness as the target class and MIS-C label and/or cluster membership as the predictors.
2.5. Features distinguishing MIS-C and severe COVID-19 respiratory illness
To identify features distinguishing patients closest to the decision boundary between MIS-C and no MIS-C labels and between clusters 1 and 2, we selected patients whose MIS-C label diverged from the majority label for the cluster in > 80% of bootstrap replications. That is, we identified patients who were not labeled as having MIS-C but consistently clustered with cluster 1 (N = 26) and patients labeled as having MIS-C who consistently clustered with cluster 2 (N = 82). We focus on these patients because they represent the patients that may be most difficult to distinguish clinically whether their disease represents MIS-C or hyperinflammation in the setting of acute COVID-19 disease. We identified key differentiating variables between these groups using backwards selection (appendix, p. 4).
2.6. Role of the funding source
The CDC participated in design and conduct of the registry; collection, management, and interpretation of data; preparation, review, and approval of the manuscript; and the decision to submit the manuscript for publication.
3. Results
As of 4 January 2021, data abstraction was completed for 1526 patients from 62 US hospitals included in the Overcoming COVID-19 registry. Of 1526 included patients, 684 (45%) were labeled by clinician investigators as having MIS-C (Table 1). Overall, 1097 (72%) tested positive by RT-PCR for SARS-CoV-2, including 327 (48%) of 684 labeled MIS-C.
Table 1.
Demographic and baseline clinical characteristics of the study cohort. All data are shown as N (%).
| Variable | Overall | MIS-C labeled | Not MIS-C labeled |
|---|---|---|---|
| N | 1526 | 684 | 842 |
| Sex (male) | 840 (55) | 398 (58) | 442 (52) |
| Age (years) | |||
| 0 – <1 | 242 (16) | 38 (6) | 204 (24) |
| 1–5 | 332 (22) | 196 (29) | 136 (16) |
| 6–12 | 428 (28) | 277 (40) | 151 (18) |
| 13–21 | 521 (34) | 172 (25) | 349 (41) |
| Missing/unknown | 3 (0) | 1 (0) | 2 (0) |
| Race/Ethnicity | |||
| White, non-Hispanic | 278 (18) | 109 (16) | 169 (20) |
| Black, non-Hispanic | 408 (27) | 213 (31) | 195 (23) |
| Hispanic or Latino | 570 (37) | 226 (33) | 344 (41) |
| Other, non-Hispanic | 100 (7) | 39 (6) | 61 (7) |
| Unknown | 170 (11) | 97 (14) | 73 (9) |
| Insurance | |||
| Public | 1018 (67) | 417 (61) | 601 (71) |
| Self-pay | 56 (4) | 30 (4) | 26 (3) |
| Private | 354 (23) | 188 (27) | 166 (20) |
| Other or Unknown | 98 (6) | 49 (7) | 49 (6) |
| At least one underlying condition* | 737 (48) | 221 (32) | 516 (61) |
| Respiratory | 302 (20) | 90 (13) | 212 (25) |
| Cardiovascular | 102 (7) | 23 (3) | 79 (9) |
| Neurological/Neuromuscular | 171 (11) | 35 (5) | 136 (16) |
| Oncologic | 55 (4) | 10 (1) | 45 (5) |
| Immunosuppressed | 40 (3) | 4 (1) | 36 (4) |
| Rheumatologic/Autoimmune | 24 (2) | 7 (1) | 17 (2) |
| Hematologic | 87 (6) | 13 (2) | 74 (9) |
| Renal or Urologic | 64 (4) | 10 (1) | 54 (6) |
| Gastrointestinal/Hepatic | 155 (10) | 27 (4) | 128 (15) |
| Endocrine | 118 (8) | 18 (3) | 100 (12) |
| Genetic/Metabolic (excluding obesity) | 63 (4) | 12 (2) | 51 (6) |
| Trisomy 21 | 13 (1) | 2 (0) | 11 (1) |
Underlying conditions are not mutually exclusive.
3.1. Identification of three clusters
A model with three clusters was optimal (appendix, p. 12) in 654 (65%) of 1000 bootstrap replications. Clusters appeared to be stable (appendix, p. 13). Among 1526 patients, there were mean (95% confidence interval [CI]) 498 (447–558) patients in cluster 1, 445 (353–667) in cluster 2, and 583 (381–667) in cluster 3 (appendix, p. 10). Feature selection yielded a model with 60 variables, including 33 clinical features and 13 biomarkers, as well as 14 indicator variables (appendix, p. 14). The variables with the strongest contribution to cluster assignment included myocarditis, lower respiratory tract infection on presentation, and treatment with supplemental oxygen or mechanical ventilation (appendix, p. 15). The selected variables accurately classified patients to clusters (90% accuracy using 5-fold cross-validation).
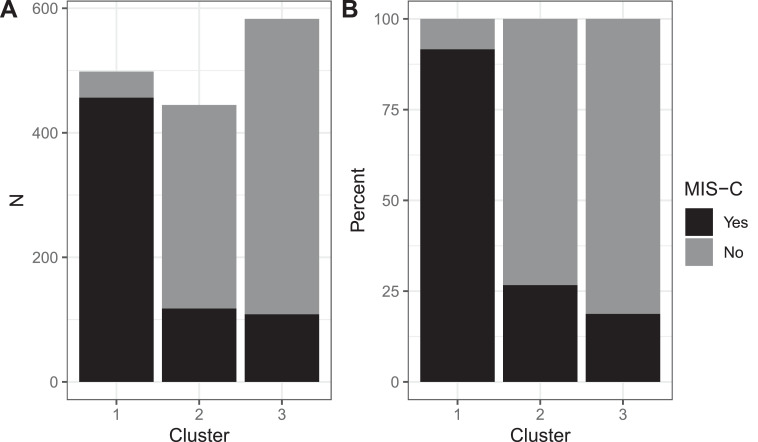
Among 684 patients labeled as having MIS-C, mean (percent ± standard deviation) 456 (67% ± 3%) were in cluster 1, 118 (17% ± 3%) were in cluster 2, and 109 (16% ± 3%) were in cluster 3 (Fig. 1A). Most of the patients (mean 456 of 498 [92% ± 2%]) in cluster 1 were labeled as having MIS-C (Fig. 1B).
Fig. 1.
Proportion of patients within each cluster with a clinical label of multisystem inflammatory system in children (MIS-C). (A) Mean number of patients by cluster. (B) Mean percentage of patients in each cluster.
3.2. Patient characteristics by cluster
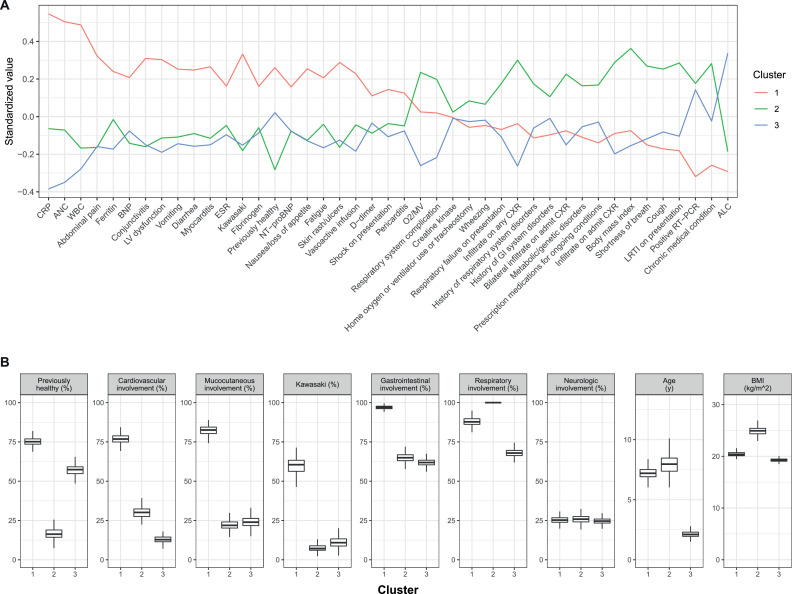
There was notable variability in demographics, signs, and symptoms among the three clusters (Fig. 2).
Fig. 2.
Overview of clinical variables among clusters. (A) Overview of differences among biomarkers and clinical features. For continuous variables, values are standardized to have mean 0 and unit standard deviation across all patients. For categorical variables, values are expressed as percent of patients with that feature, standardized to have mean 0 across all patients; thus, standardized values represent the proportion in each cluster above or below the overall mean proportion of patients having that feature. (B) Demographic and general clinical features among patients by cluster. ALC = absolute lymphocyte count; ANC = absolute neutrophil count; BMI = body mass index; BNP = B-type natriuretic peptide; CK = creatine kinase; CRP = C-reactive protein; ESR = erythrocyte sedimentation rate; MV = mechanical ventilation; NT-proBNP = N-terminal pro b-type natriuretic peptide; O2 = oxygen; WBC = white blood cell count.
Demographics: Patients in clusters 1 and 2 were older (mean 7·2 ± 0·4 y and 7·4 ± 2·1 y, respectively) compared with those in cluster 3 (2·8 ± 2·0 y). Mean BMI in cluster 2 (24·3 ± 2·1) was higher than in clusters 1 (20·5 ± 0·45) and 3 (19·9 ± 1·9). Most patients (mean 75% ± 3%) in cluster 1 were previously healthy, whereas 79% ± 15% of patients in cluster 2 and 49% ± 15% in cluster 3 had at least one underlying medical condition (appendix, p. 10).
Biomarkers and SARS-CoV-2 testing: More patients in clusters 2 (90% ± 4%) and 3 (86% ± 3%) had positive RT-PCR testing for SARS-CoV-2 than patients in cluster 1 (40% ± 3%). More patients in cluster 1 (81% ± 2% vs. 19% ± 3% in cluster 2 and 19% ± 2·9% in cluster 3) had positive testing for SARS-CoV-2 antibodies (Fig. 3A). This pattern of differences persisted when grouping patients in each cluster by whether they were labeled as having MIS-C, with 75% ± 10% of MIS-C-labeled patients in cluster 2 having positive RT-PCR results versus only 40% ± 2% of MIS-C-labeled patients in cluster 1 (appendix, p. 16). Compared with clusters 2 and 3, patients in cluster 1 had higher inflammatory marker levels, including white blood cell (WBC) count, neutrophil to lymphocyte ratio, C-reactive protein (CRP), erythrocyte sedimentation rate (ESR) and fibrinogen, and higher D-dimers (Fig. 3B). Cluster 1 patients also had higher B-type natriuretic peptide (BNP) levels. These biomarkers were higher in MIS-C-labeled patients compared with those not labeled as having MIS-C in all clusters, although levels among MIS-C labeled patients differed across clusters, particularly for WBC count, platelets, CRP, and BNP (appendix, p. 16).
Fig. 3.
Comparison of features by cluster. Boxplots show median (heavy horizontal line), 25th and 75th percentiles (bounds of box), and 150% the interquartile range below and above the 25th and 75th percentiles (vertical lines). (A) Clinical features. (B) Biomarkers. PARDS = pediatric acute respiratory distress syndrome.
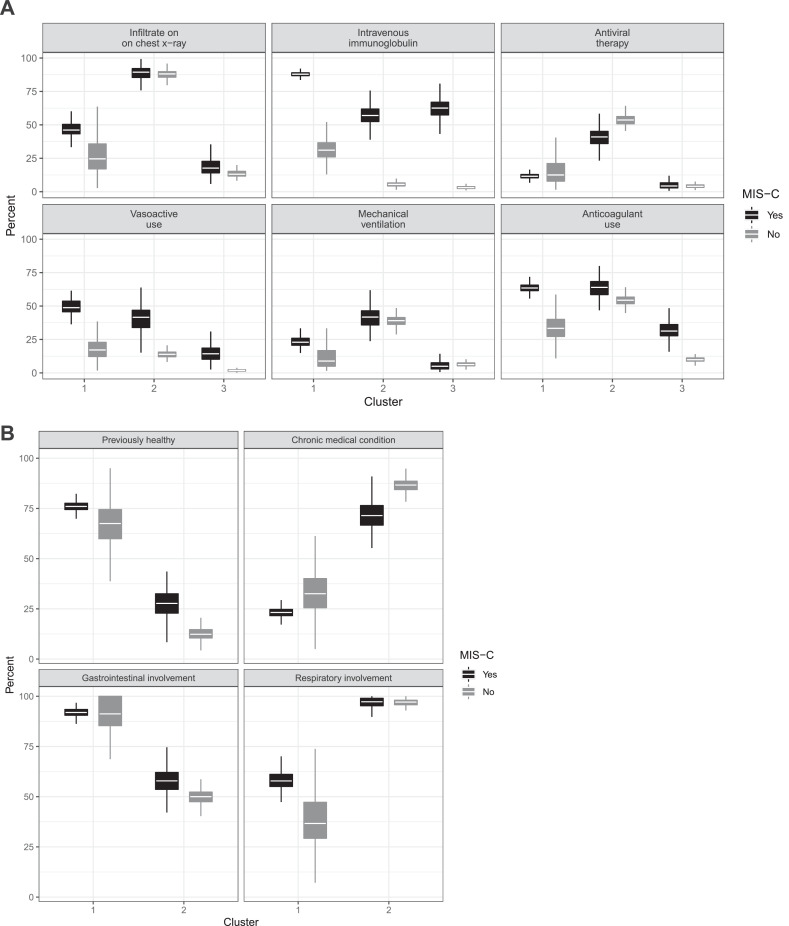
Clinical signs and symptoms: More patients in cluster 1 had mucocutaneous involvement and Kawasaki-like disease on presentation (Fig. 2). Cardiovascular involvement was nearly three times as common for patients in cluster 1 compared with patients in clusters 2 and 3 (77% ± 3% vs. 29% ± 6% vs. 14% ± 5%). Coronary artery aneurysms were infrequent in all clusters, though they were more commonly observed in cluster 1 (14% ± 2%) than in clusters 2 (4% ± 1%) and 3 (2% ± 0·8%). Although most patients in all clusters had some respiratory system involvement, 22% ± 8% of patients in cluster 2 met criteria for pediatric ARDS (PARDS), compared with 7% ± 2% of patients in cluster 1 and 4% ± 8% of patients in cluster 3 (Fig. 3A). Nearly all patients in cluster 2—both those labeled and not labeled as having MIS-C—had infiltrates on chest radiography during hospitalization, whereas only a minority of patients in clusters 1 and 3 had this finding (Figs. 3A and 4A).
Fig. 4.
Characteristics differentiating MIS-C-labeled and MIS-C non-labeled patients. Boxplots show median (white horizontal line), 25th and 75th percentiles (bounds of box), and 150% the interquartile range below and above the 25th and 75th percentiles (vertical lines). (A) MIS-C patients in cluster 2 had similar prevalence of infiltrates on chest x-rays, mechanical ventilation, and treatment with antiviral therapy as non-MIS-C patients. However, MIS-C patients in cluster 2 also received intravenous immunoglobulin and vasoactive infusions at the same frequency as MIS-C patients in cluster 1. (B) Clinical characteristics distinguishing patients in clusters 1 and 2 were similar among MIS-C labeled and non-labeled patients in each cluster and accurately distinguished cluster 1 non-MIS-C patients from cluster 2 MIS-C labeled patients. Gastrointestinal involvement includes abdominal pain, diarrhea, and vomiting. Respiratory involvement includes presence of infiltrates on chest radiographs, presentation with lower respiratory tract infection, and cough.
Treatments: Patients in cluster 1 were more likely overall to receive immunomodulatory therapy (89% ± 2% vs. 44% ± 8% in cluster 2 and 22% ± 9% in cluster 3), including intravenous immunoglobulin (IVIg) (Fig. 3A). However, patients labeled as having MIS-C were more likely to be treated with IVIg, regardless of cluster (Fig. 4A). In contrast, patients in cluster 2 were more likely to be treated with SARS-CoV-2 antiviral therapy whether labeled as having MIS-C or not (Fig. 4A). Anticoagulant use frequency was greater in clusters 1 and 2 (Fig. 3A) and was similarly frequent in cluster 2 regardless of MIS-C label (Fig. 4A). Systemic corticosteroids were also more frequently administered to patients in cluster 1 (70% ± 3%) than to those in cluster 2 (49% ± 12%), and this difference persisted regardless of MIS-C label. Of note, the reason for treatment with corticosteroids in 24% of patients treated with steroids and not labeled ultimately as having MIS-C was nevertheless “suspected MIS-C”; these patients were typically in cluster 2.
3.3. Association between clusters and critical illness outcomes
Patients in clusters 1 and 2 had similar frequency of intensive care unit admission (75% ± 3 vs. 67% ± 14%), whereas the frequency was lower in cluster 3 (36% ± 13%). Critical illness outcomes were more common in patients in cluster 1 (53% ± 5%) and cluster 2 (44% ± 12%) than in cluster 3 (17% ± 11%). However, patterns of severe outcomes differed between clusters 1 and 2. Need for invasive or non-invasive mechanical ventilation was more common in cluster 2 (36% ± 12%) compared with cluster 1 (22% ± 4%). In contrast, vasoactive use was more common in cluster 1 (47% ± 5%) than in cluster 2 (20% ± 7%). Death was rare in all clusters, but more patients in cluster 2 died (3% ± 1%) compared to clusters 1 (0·8% ± 0·5%) and 3 (1% ± 1%).
By multivariable regression, cluster assignment and MIS-C label both independently discriminated critical illness, with C-statistic 0·73 ± 0·02. However, by all three metrics analyzed and in all 1000 bootstrap replications, cluster assignment contributed more than MIS-C label to models’ classification of a patient's having critical illness. Models with cluster membership alone as compared to MIS-C label alone had better discrimination (C-statistic 0·71 ± 0·03 vs. 0·64 ± 0·01), and p-values for models with cluster membership alone were lower than those with MIS-C label alone. Cluster membership was also the more important variable by dominance analysis. Using recursive feature elimination and a random forest model, cluster membership was the more important variable in 663 of 1000 replications.
3.4. Features distinguishing MIS-C from severe COVID-19 respiratory illness
Patients in cluster 1 not labeled as having MIS-C were distinguished from those in cluster 2 labeled as having MIS-C by presence or absence of underlying conditions and by gastrointestinal and respiratory system involvement. Regardless of MIS-C label, patients in cluster 1 were more often previously healthy and had more frequent gastrointestinal system involvement, whereas patients in cluster 2 had prior medical conditions and more frequent respiratory system involvement (Fig. 4B). Using these variables in a random forest model differentiated cluster 1 patients not labeled as having MIS-C from those in cluster 2 labeled as having MIS-C with 97% accuracy. The most important variable distinguishing these patients, whose MIS-C status was potentially mislabeled, was presence of infiltrates on chest radiographs (appendix, p. 17).
4. Discussion
MIS-C criteria are broad and encompass other critical illness syndromes such as PARDS with secondary organ involvement [5,6,8,26]. We identified three clusters of phenotypically distinct patients among a large cohort of patients hospitalized with COVID-19-related illness. Cluster 1, which we subsequently refer to as the “prototypical MIS-C” cluster, included previously healthy patients with cardiovascular and/or mucocutaneous involvement, often presenting with gastrointestinal symptoms, who had marked elevations of inflammatory markers and BNP. Many had SARS-CoV-2 test results that appear post-infectious (i.e., antibody positive but PCR negative) [5,27]. Indeed, 92% of patients in the prototypical MIS-C cluster were labeled by clinicians and clinical researchers as having MIS-C. Cluster 2, or the “respiratory cluster”, were mostly SARS-CoV-2 PCR positive, had chronic conditions (most commonly respiratory), had infiltrates on chest radiographs, and many needed mechanical ventilator support and had a diagnosis of PARDS. Nearly 20% of patients diagnosed with MIS-C by clinicians were in cluster 2, including many that received vasoactive agents. Patients in cluster 3 were younger and less critically ill, and of the 16% of MIS-C-labeled patients in that cluster, 44% had KD features. It is possible that patients labeled as MIS-C that were not in the prototypical MIS-C cluster could be a subset with distinct phenotypes.
A prior study by Godfred-Cato and colleagues applied latent class analysis to cluster patients with MIS-C, identifying three subphenotypes [14]. They also identified a respiratory cluster among patients with MIS-C, noting they likely had acute COVID-19, but did not include other acute COVID-19 patients in their analysis for comparison. By including patients not labeled by clinicians as having MIS-C, we confirm that this subgroup of MIS-C is phenotypically similar to patients known to have acute COVID-19 lung disease. The Godfred-Cato study reported two other subphenotypes-one with cardiovascular and gastrointestinal involvement and one with KD-like features. In contrast, our analysis revealed one highly inflamed cluster that predominantly included both patients with cardiovascular involvement and those with KD features. Our approach was similar to latent class analysis of adult ARDS patients that identified a hyperinflammatory subphenotype associated with differences in treatment response [28].
We previously reported that most patients with MIS-C have respiratory involvement similar to patients with acute COVID-19 [26], and therefore its inclusion in the criteria for MIS-C may decrease diagnostic specificity. The current work builds on the prior findings by showing empirically that a subset of patients labeled as MIS-C appear phenotypically more similar to COVID-19 patients in the respiratory cluster. We identified a novel, important variable distinguishing respiratory COVID-19 disease from MIS-C, namely, pulmonary infiltrates on chest radiographs. Respiratory involvement is not included as part of the multiorgan involvement criteria of the World Health Organization (WHO) case definition for MIS-C, but is allowed in the CDC criteria for organ involvement [12,13]. Clinicians should consider whether pulmonary infiltrates, especially non-cardiogenic pulmonary edema, may suggest acute COVID-19 infection rather than MIS-C. Importantly, our data suggest that many of these patients are treated with SARS-CoV-2-targeted antiviral therapy despite being labeled as having MIS-C and being treated with IVIg for that disorder. Conversely, many patients in the respiratory cluster, ultimately determined not to have MIS-C, were nonetheless treated with systemic corticosteroids reportedly with MIS-C as the indication. This treatment approach suggests that when clinicians cannot accurately distinguish between MIS-C and acute COVID-19 pneumonia, they are treating both phenotypes simultaneously, and better delineation between the two diagnoses may allow more precise treatment.
Cardiovascular involvement and hyperinflammation are predominant features of MIS-C. Although lymphopenia was common in both the prototypical MIS-C cluster and the respiratory cluster, prototypical MIS-C patients were more likely to have neutrophilia, higher CRP, and higher BNP. Standardizing the use of diagnostic screening tests, such as complete blood count with differential, CRP, and BNP, could help to develop cutoffs that distinguish acute COVID-19 from MIS-C.
Our work had several limitations. Laboratory values were measured based on clinical judgment and missingness was likely non-random. We therefore included indicator variables to allow assigning a mean value to the missing laboratory variable without biasing the model. More refined clusters may become evident by incorporating temporal trends in laboratory values. We lacked accurate data on timing between COVID-19 exposure or infection with SARS-CoV-2 and hospitalization. Although prevalence of RT-PCR positivity and antibody response may suggest inferences regarding MIS-C as a post-infectious phenomenon distinct from acute COVID-19 [29,30], we cannot directly test this hypothesis. Lack of data prevented using timing of exposure to analyze association of laboratory values with disease progression. Additional variables not included in our models may also help refine clusters. We also did not have a large, independent cohort to validate our clustering results. However, our approach does not train a classifier, so any potential biases are inherent to the population studied rather than to the algorithm itself. Although external validation of performance is not mandatory for clustering analyses, whether patients from different populations exhibit similar clustering patterns merits confirmation. Whether appropriate MIS-C labeling and treatment decisions affect patient outcomes remains unknown.
Applying an unsupervised, data driven analysis to children and adolescents hospitalized with the full spectrum of COVID-19-associated complications, we identified a cluster of features characteristic of patients with prototypical MIS-C and another cluster with likely respiratory complications of severe COVID-19. The clustering analysis further identified patients who were labeled by clinicians as having MIS-C but who may have distinct subphenotypes, including acute COVID-19 with cardiovascular involvement in the respiratory cluster or Kawasaki disease in the less ill cluster. The extent to which these subphenotypes should be included in the broader MIS-C definition or represent distinct disease entities merits further study. Our findings may be helpful in refining the criteria for MIS-C to identify phenotypes that have different pathophysiology and that possibly need alternate therapeutic strategies. Clinicians should especially question whether patients with preexisting conditions and non-cardiogenic pulmonary edema may have acute COVID-19 infection rather than MIS-C and tailor treatment accordingly.
Contributors
AG, MMP, KDM, LRF, MWT, JWN, and AGR conceived and designed the study. AG, MMN, CCY, LRF, MWT, and AGR performed the data curation. AG, KDM, and AGR performed the formal analysis. MMP, MMN, and AGR acquired the funding for the current study. All authors contributed to the investigation and data collection. AG, MMP, KDM, and AGR designed the methodology. All authors contributed to the project administration. MMN, MDL, and AGR provided the resources to perform the study. AG and CCY designed the software used in the study. MMP and AGR supervised the study. AG and MMP validated the findings. AG created the data visualizations. AG, MMP, KDM, and AGR drafted the manuscript. All authors made important intellectual contributions to the interpretation of data and helped revise the manuscript. AG, CCY, and AGR have verified the underlying data. AG, MMN, CCY, and AGR accessed and were responsible for the raw data associated with the study. All site authors had full access to the data from their sites in the study and all authors accept responsibility to submit for publication.
Declaration of Competing Interest
All authors report receiving funding from the Centers for Disease Control and Prevention for the current study. AG reports receiving grants from the NIH outside of the submitted work. ABM reports receiving grants from the Francis Family Foundation and from the NIH/NICHD (K23HD096018) outside of the submitted work. CVH reports receiving consulting fees from DYNAMED and BIOFIRE outside of the submitted work. JES reports receiving grants from Merck outside of the submitted work. NBH reports receiving grants from Sanofi, Quidel, the NIH, and the CDC; consulting fees from Moderna; and an educational grant from Genetech outside of the submitted work. CMR reports receiving grants from the NIH/NHLBI (K23HL150244) outside of the submitted work. NZC reports receiving grants or contracts from Boston Children's Hospital and Cincinnati Children's Hospital and Medical Center outside of the submitted work. JCF reports receiving grants from the NIH outside of the submitted work. HD reports payments from Delex Pharma International Inc. outside of the submitted work. PMM reports receiving grants from the NIH and serving as a member of data safety monitoring board for the NIH supported KIDS-DOT trial outside of the submitted work. AGR reports receiving royalties from UpToDate outside of the submitted work. All other authors have nothing to declare.
Acknowledgments
Data sharing statement
The data dictionary defining each field in the dataset used in this analysis, the study protocol, and R code used in the analyses are available upon request. Access to a deidentified version of the dataset, including individual participant data, is possible through collaboration with the investigators after approval of a proposal and data use approval of participating sites.
Acknowledgements
We appreciate and thank the many research coordinators at the Overcoming COVID-19 hospitals who assisted in data collection for this study. We thank the leadership of the Pediatric Acute Lung Injury and Sepsis Investigator's (PALISI) Network for their ongoing support.
Funding
This work was funded by the US Centers for Disease Control and Prevention (75D30120C07725) and National Institutes of Health (K12HD047349 and R21HD095228).
Disclaimer
The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.eclinm.2021.101112.
Appendix. Supplementary materials
References
- 1.Verdoni L., Mazza A., Gervasoni A. An outbreak of severe Kawasaki-like disease at the Italian epicentre of the SARS-CoV-2 epidemic: an observational cohort study. Lancet. 2020;395:1771–1778. doi: 10.1016/S0140-6736(20)31103-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Toubiana J., Poirault C., Corsia A. Kawasaki-like multisystem inflammatory syndrome in children during the covid-19 pandemic in Paris, France: prospective observational study. BMJ. 2020;369:m2094. doi: 10.1136/bmj.m2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pouletty M., Borocco C., Ouldali N. Paediatric multisystem inflammatory syndrome temporally associated with SARS-CoV-2 mimicking Kawasaki disease (Kawa-COVID-19): a multicentre cohort. Ann Rheum Dis. 2020;79:999–1006. doi: 10.1136/annrheumdis-2020-217960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Riphagen S., Gomez X., Gonzalez-Martinez C., Wilkinson N., Theocharis P. Hyperinflammatory shock in children during COVID-19 pandemic. Lancet. 2020;395:1607–1608. doi: 10.1016/S0140-6736(20)31094-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Feldstein L.R., Rose E.B., Horwitz S.M. Multisystem inflammatory syndrome in U.S. children and adolescents. N Engl J Med. 2020;383:334–346. doi: 10.1056/NEJMoa2021680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Whittaker E., Bamford A., Kenny J. Clinical characteristics of 58 children with a pediatric inflammatory multisystem syndrome temporally associated with SARS-CoV-2. JAMA. 2020;324:259–269. doi: 10.1001/jama.2020.10369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dufort E.M., Koumans E.H., Chow E.J. Multisystem inflammatory syndrome in children in New York state. N Engl J Med. 2020;383:347–358. doi: 10.1056/NEJMoa2021756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Belot A., Antona D., Renolleau S. SARS-CoV-2-related paediatric inflammatory multisystem syndrome, an epidemiological study, France, 1 March to 17 May 2020. Euro Surveill. 2020;25 doi: 10.2807/1560-7917.ES.2020.25.22.2001010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheung E.W., Zachariah P., Gorelik M. Multisystem inflammatory syndrome related to COVID-19 in previously healthy children and adolescents in New York city. JAMA. 2020;324:294–296. doi: 10.1001/jama.2020.10374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.European Centre for Disease Prevention and Control. Paediatric inflammatory multisystem syndrome and SARS-CoV-2 infection in children [Internet]. Stockholm: European Centre for Disease Prevention and Control; 15 May 2020 [cited 27 Oct 2020]. Available from: https://www.ecdc.europa.eu/sites/default/files/documents/covid-19-risk-assessment-paediatric-inflammatory-multisystem-syndrome-15-May-2020.pdf
- 11.Royal College of Paediatrics and Child Health. Guidance: paediatric multisystem inflammatory syndrome temporally associated with COVID-19 [Internet]. London: Royal College of Paediatrics and Child Health; 01 May 2020 [cited 27 Oct 2020]. Available from: https://www.rcpch.ac.uk/sites/default/files/2020-05/COVID-19-Paediatric-multisystem-%20inflammatory%20syndrome-20200501.pdf
- 12.Centers for Disease Control and Prevention. Multisystem Inflammatory Syndrome in Children (MIS-C) Associated with Coronavirus Disease 2019 (COVID-19) [Internet]. Atlanta: Centers for Disease Control and Prevention; 14 May 2020 [cited 30 Sep 2020]. Available from: https://emergency.cdc.gov/han/2020/han00432.asp
- 13.World Health Organization. Multisystem inflammatory syndrome in children and adolescents temporally related to COVID-19 [Internet]. Geneva: World Health Organization; 15 May 2020 [updated 17 May 2020; cited 30 Sep 2020]. Available from: https://www.who.int/news-room/commentaries/detail/multisystem-inflammatory-syndrome-in-children-and-adolescents-with-covid-19
- 14.Godfred-Cato S., Bryant B., Leung J. COVID-19-associated multisystem inflammatory syndrome in children-United States, March-July 2020. MMWR Morb Mortal Wkly Rep. 2020;69:1074–1080. doi: 10.15585/mmwr.mm6932e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Consiglio C.R., Cotugno N., Sardh F. The immunology of multisystem inflammatory syndrome in children with COVID-19. Cell. 2020 doi: 10.1016/j.cell.2020.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim L., Whitaker M., O'Halloran A. Hospitalization rates and characteristics of children aged <18 years hospitalized with laboratory-confirmed COVID-19 -COVID-NET, 14 States, March 1-July 25, 2020. MMWR Morb Mortal Wkly Rep. 2020;69:1081–1088. doi: 10.15585/mmwr.mm6932e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gotzinger F., Santiago-Garcia B., Noguera-Julian A. COVID-19 in children and adolescents in Europe: a multinational, multicentre cohort study. Lancet Child Adolesc Health. 2020;4:653–661. doi: 10.1016/S2352-4642(20)30177-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Swann O.V., Holden K.A., Turtle L. Clinical characteristics of children and young people admitted to hospital with COVID-19 in United Kingdom: prospective multicentre observational cohort study. BMJ. 2020;370:m3249. doi: 10.1136/bmj.m3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matthay M.A., Zemans R.L., Zimmerman G.A. Acute respiratory distress syndrome. Nat Rev Dis Prim. 2019;5:18. doi: 10.1038/s41572-019-0069-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Most Z.M., Hendren N., Drazner M.H., Perl T.M. Striking similarities of multisystem inflammatory syndrome in children and a myocarditis-like syndrome in adults: overlapping manifestations of COVID-19. Circulation. 2021;143:4–6. doi: 10.1161/CIRCULATIONAHA.120.050166. [DOI] [PubMed] [Google Scholar]
- 21.von Elm E., Altman D.G., Egger M. The strengthening the reporting of observational studies in epidemiology (STROBE) statement: guidelines for reporting observational studies. Lancet. 2007;370:1453–1457. doi: 10.1016/S0140-6736(07)61602-X. [DOI] [PubMed] [Google Scholar]
- 22.Apple F.S., Collinson P.O. Biomarkers ITFoCAoC. Analytical characteristics of high-sensitivity cardiac troponin assays. Clin Chem. 2012;58:54–61. doi: 10.1373/clinchem.2011.165795. [DOI] [PubMed] [Google Scholar]
- 23.Gao L.L., Bien J., Witten D. Selective inference for hierarchical clustering. arXiv:2012.02936 [Preprint]. 5 Dec 2020 [cited 21 Jul 2021]. Available from: https://arxiv.org/abs/2012.02936
- 24.Azen R., Traxel N. Using dominance analysis to determine predictor importance in logistic regression. J Educ Behav Stat. 2009;34:319–347. [Google Scholar]
- 25.Nagelkerke N.J.D. A note on a general definition of the coefficient of determination. Biometrika. 1991;78:691–692. [Google Scholar]
- 26.Feldstein L.R., Tenforde M.W., Friedman K.G. Characteristics and outcomes of US children and adolescents with multisystem inflammatory syndrome in children (MIS-C) compared with severe acute COVID-19. JAMA. 2021;325:1074–1087. doi: 10.1001/jama.2021.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Belay E.D., Abrams J., Oster M.E. Trends in geographic and temporal distribution of US children with multisystem inflammatory syndrome during the COVID-19 pandemic. JAMA Pediatr. 2021 doi: 10.1001/jamapediatrics.2021.0630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Calfee C.S., Delucchi K., Parsons P.E. Subphenotypes in acute respiratory distress syndrome: latent class analysis of data from two randomised controlled trials. Lancet Respir Med. 2014;2:611–620. doi: 10.1016/S2213-2600(14)70097-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Long Q.X., Liu B.Z., Deng H.J. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat Med. 2020;26:845–848. doi: 10.1038/s41591-020-0897-1. [DOI] [PubMed] [Google Scholar]
- 30.Azkur A.K., Akdis M., Azkur D. Immune response to SARS-CoV-2 and mechanisms of immunopathological changes in COVID-19. Allergy. 2020;75:1564–1581. doi: 10.1111/all.14364. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.