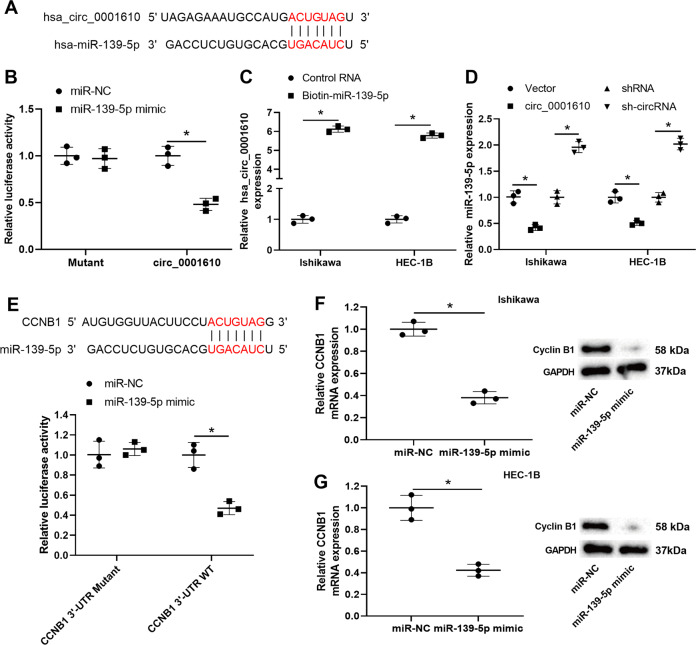
Fig. 4. The interplay between hsa_circ_0001610, microRNA (miR)-139-5p, and cyclin B1.
A The potential binding sites between hsa_circ_0001610 and miR-139-5p. B Dual-luciferase reporter assay using HEK-293T cells co-transfected with pmirGLO-circ_0001610 vector/pmirGLO-mutant-circ_0001610 vector and miR-139-5p mimic/the negative control of miR-139-5p mimic (miR-NC). C qRT-PCR analysis of hsa_circ_0001610 level in the streptavidin captured fractions from the Ishikawa and HEC-1B cell lysates after incubation with biotinylated miR-139-5p (biotin-miR-139-5p) or control RNA. D qRT-PCR analysis of miR-139-5p in the Ishikawa and HEC-1B cells after transfection with circ_0001610/the negative control of circ_0001610 (vector)/sh-circRNA/shRNA. E Upper: the potential binding sites between CCNB1 (the encoding gene of cyclin B1) and miR-139-5p. Below: dual-luciferase reporter assay using HEK-293T cells co-transfected with pmirGLO-CCNB1 3′-UTR mutant vector/ pmirGLO-CCNB1 3′-UTR wild type (WT) vector and miR-139-5p mimic/ miR-NC. The mRNA and protein levels of cyclin B1 were measured in F Ishikawa and G HEC-1B cells after transfection with miR-139-5p mimic/miR-NC. *p < 0.05.