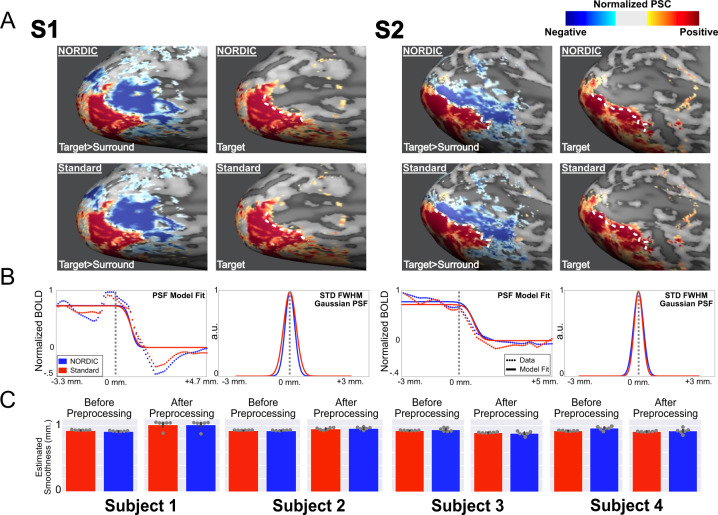
Fig. 5. Functional point spread function (PSF) and global image smoothness.
Panel A, top row shows NORDIC normalized beta percent signal change (PSC) maps for differential mapping target (in red) > surround (in blue) (left), and the target only (right) single-condition image for subjects 1 and 2. The white dotted line is determined in the differential image as the “boundary” between the two stimulations. The same white dotted line is also superimposed on the target-only PSC map where PSC values are greater than zero but decreasing in magnitude progressively away from this “boundary” posteriorly. The functional PSF is calculated from this spread in PSC beyond the “boundary”. Panel A, the lower row is identical to the upper row but obtained from Standard reconstruction data. Panel B left panel for each subject: the PSC magnitude changes (normalized to the highest value) along traces perpendicular to the “boundary” are displayed as the average (across traces and runs). The model fits (solid line) and data (dotted line) are shown for both the NORDIC (blue) and Standard (red) reconstructions. The vertical gray dotted line represents the “boundary” as derived from the differential maps. Panel B, right panel for each subject portrays the full-width at half maximum (FWHM) standard deviation of the gaussian kernel that was convolved with a step function to model functional PSF (see the “Methods” section). Panel C: Mean global smoothness of images used for the fMRI time series for Standard (red) and NORDIC (blue) in four subjects, before (left panel) and after preprocessing related interpolations. Error bars represent the standard error of the mean across 6 independent runs (shown as gray dots). Source data are provided as a source Data file.