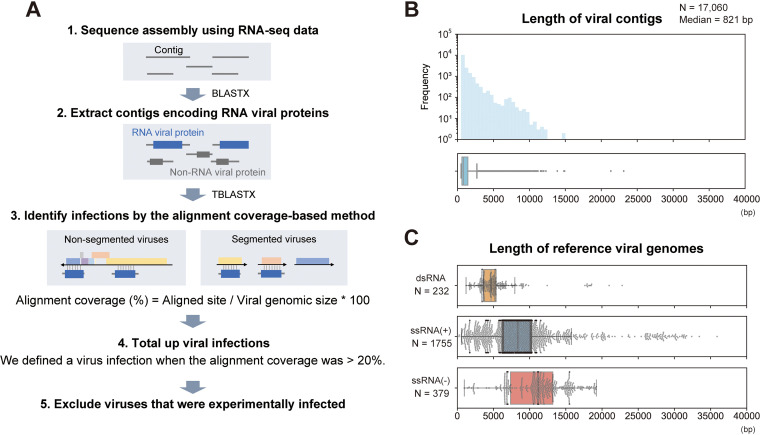
FIG 1.
Strategy for detecting viral infections in public RNA-seq data. (A) Schematic diagram of the procedure for detecting viral infections. First, we performed de novo sequence assembly using publicly available mammalian and avian RNA-seq data. Second, we extracted contigs encoding RNA viral proteins by BLASTX. Third, we constructed sequence alignments by TBLASTX using the viral contigs in each RNA-seq data and reference viral genomes because most viral contigs were shorter than complete viral genomes, as shown in panels B and C. The alignment coverage is defined as the proportion of aligned sites in the entire reference viral genome. Fourth, we determined a viral infection when the alignment coverage was >20%. Finally, we totaled the infections at the virus family level after excluding experimentally infected viruses (see Materials and Methods). (B) Distributions of viral contig length: histogram (upper panel) and box plot (lower panel). The x axis indicates the viral contig length. Among 17,060 viral contigs, the median length was 821 bp. (C) Length of reference viral genomes. Each panel corresponds to the Baltimore classification: the upper, middle, and lower panels show double-stranded RNA (dsRNA) viruses, positive-sense single-stranded RNA [ssRNA(+)] viruses, and negative-sense single-stranded RNA [ssRNA(−)] viruses, respectively. The x axis indicates the viral genome size. These viral genomes were obtained from the RefSeq genomic viral database. The genomic size of segmented viruses is the sum length of all segments in a virus species.